- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Viral Double-Stranded RNA Detection by DNase I and Nuclease S1 digestions in Leishmania parasites
Published: Vol 10, Iss 9, May 5, 2020 DOI: 10.21769/BioProtoc.3598 Views: 5888
Reviewed by: Alexandros AlexandratosAmit DeyOmar Akil

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Direct RNA Sequencing of Foot-and-mouth Disease Virus Genome Using a Flongle on MinION
Lizhe Xu [...] Bonto Faburay
Jun 20, 2024 2144 Views

Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay
Yidan Sun [...] Chao Shen
Dec 5, 2024 1618 Views

Novel Workflows for Separate Isolation of Pathogen RNA or DNA From Wastewater: Detection by Innovative and Conventional qPCR
Kristina M. Babler [...] Ayaaz Amirali
Feb 20, 2025 3196 Views
Abstract
Many RNA viruses are found in protozoan parasites. They can be responsible for more serious pathology or treatment failure. For the detection of viral double-stranded RNA (dsRNA), sequence-dependent and -independent methods are available, such as quantitative real-time PCR and immunofluorescence, dot blot, ELISA or sequencing. The technique presented here is sequence-independent and is well detailed in the following protocol, taking the example of Leishmania RNA virus (LRV) in Leishmania guyanensis (Lgy) species. To summarise, the protocol is divided into four major steps: RNA extraction from the parasites, RNA purification, enzymatic digestions with DNase I and Nuclease S1, and visualization by gel electrophoresis. This method can be used to detect other viral dsRNA in other parasites. It provides an additional tool, complementary to other techniques previously cited and it is easy and quite fast to achieve.
Keywords: dsRNABackground
The wide diversity of RNA viruses present in protozoan parasites has been well documented (Wang and Wang, 1991; Ghosh et al., 2012; Zangger et al., 2014; Lye et al., 2016; Akopyants et al., 2016; Fernandez-Presas et al., 2017; Grybchuk et al., 2018). Moreover, these viruses have been described as potential virulence factors (Fichorova et al., 2013; El-Gayar et al., 2016; Rath et al., 2019). Of particular note, the presence of the endosymbiont Leishmania RNA virus (LRV), a Totiviridae double-stranded RNA (dsRNA) virus, in Leishmania guyanensis (Lgy) exacerbates leishmaniasis disease (Ives et al., 2011; Rossi et al., 2017), favors metastasis by inducing interleukin-17 (Hartley et al., 2016) and also increases the risk of treatment failure (Adaui et al., 2016; Bourreau et al., 2016; Vieira-Gonçalves et al., 2019). This shows the importance of viral dsRNA detection in parasites. The technique presented here is sequence-independent, in comparison to quantitative real-time PCR, so it can be applied widely to RNA viruses. This dsRNA detection protocol can also, and is recommended, to be used to confirm (or be confirmed with) other methods of detection that are sequence-independent or -dependent, as for example dot blot or PCR (Zangger et al., 2013). The technique is presented here with LRV, however its application is possible for other RNA viruses and in other parasites (Grybchuk et al., 2018).
Materials and Reagents
- Eco Nitrile PF 250 Gloves (ecoSHIELDTM)
- Filter tips Low retention 1,000 µl, 200 µl, 20 µl (ClearLine, catalog numbers: 713118 , 713117 , 713115 , respectively)
- 10 µl Extended Length Filter Tip (Neptune Scientific, catalog number: BT10XL )
- Tissue culture flask 25 version “Vent” (TPP®, catalog number: 90025 )
- Vacuum Filtration 500 “rapid”-Filtermax, PES membrane 0.22 μm pore size (TPP®, catalog number: 99500 )
- Microcentrifuge 1.5 ml tubes (Corning, Axygen®, catalog number: MCT-175-C )
- Polypropylene conical 50 ml centrifuge tubes (TPP Techno Plastic Products, catalog number: 91050 )
- 2 ml serological pipettes (FALCON®, catalog number: 357507 )
- 5 ml serological pipettes (SARSTEDT, catalog number: 86.1253.001 )
- 10 ml serological pipettes (SARSTEDT, catalog number: 86.1254.001 )
- 25 ml serological pipettes (SARSTEDT, catalog number: 86.1685.001 )
- Dulbecco's Phosphate-Buffered Saline (DPBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14040091 ) stored at 4 °C
- Schneider’s Drosophila Medium w: L-Glutamine and 0.40 g/L NaHCO3 (PANTM BIOTECH, catalog number: P04-91500 ) stored at 4 °C
- Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 ), sent at -20 °C, then heat-inactivated (see Recipes from Bio-protocol by Reverte and Fasel, 2019), aliquoted to 50 ml tubes and stored at -20 °C
- HEPES Buffer 1 M (BioConcept, catalog number: 5-31F00-H ) stored at 4 °C
- Penicillin-Streptomycin (P/S) solution (10,000 IU/ml P and 10 mg/ml S) (Bioconcept, catalog number: 4-01F00-H ), sent at -20 °C, then stored at 4 °C for short-term storage
- Hemin BioXtra, from Porcine (Sigma-Aldrich, catalog number: 51280 ) stored at 4 °C
- Folic acid (FlukaTM, catalog number: 47620 ) stored at 4 °C
- 6-Biopterin (Sigma-Aldrich, catalog number: B2517 ) stored at -20 °C
- TRI® reagent (Molecular Research Center, MRC, catalog number: TR118 , 500 ml) stored at 4 °C
- Chloroform (Thermo Fisher Scientific, Fisher Chemical, catalog number: C/4960/15 ) stored at room temperature
- Isopropanol (Thermo Fisher Scientific, Fisher Chemical, catalog number: P/7490/17 ) stored at room temperature
- Ethanol absolute (Thermo Fisher Scientific, Fisher Chemical, catalog number: E/0650DF/17 ) stored at room temperature
- UltraPureTM DNase/RNase-free Distilled Water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977-035 ), shipped at room temperature, stored at 4 °C before opening, aliquoted to 1.5 ml tubes and stored at -20 °C
- DNase I, RNase-free 10 U/µl with an activity of 0.2 U/µg on calf thymus DNA (Roche, catalog number: 10 776 785 001 ) stored at -20 °C
- Nuclease S1 (#E576) (Promega, catalog number: M5761 ) stored at -20 °C
Note: Pay attention to the fact that the concentration is specific to each tube and is indicated on the tube by the manufacturer. - Buffer S1 (#E578) (Promega, catalog number: M577A ) stored at -20 °C
Note: The old reference was at 7.4x, but now Nuclease S1 Reaction Buffer is provided at 10x. - 1 kb DNA Ladder 500 µg/ml (NEB, catalog number: N3232S ) stored at 4 °C
- 6x Gel Loading Dye (Purple), no SDS (NEB, catalog number: B7025 ) stored at 4 °C; supplied with the 1 kb DNA Ladder
- Standard Agarose–Type LE for routine gel electrophoresis (BioConcept Ltd, Amimed, catalog number: 7-01P02-R ) stored at room temperature
- Tris(hydroxymethyl)aminomethane, Molecular biology (Biosolve, catalog number: 20092391 ) stored at room temperature
- Boric Acid (Crystalline Powder/Electrophoresis grade) (Fisher BioReagents, catalog number: BP168-1 ) stored at room temperature
- EDTA, Ethylenediaminetetraacetic Acid, Disodium Salt Dihydrate (Crystalline Powder/Electrophoresis grade) (Fisher BioReagents, catalog number: BP120-1 ) stored at room temperature
- SDS, Sodium Dodecylsulfate, Solution 20% pure (AppliChem PanReac, ITW Reagents, catalog number: A3942,1000 ) stored at room temperature
- Ultra-pure water, type Ultra ClearTM (BLANC-LABO) stored at room temperature
- RNase AWAYTM (Molecular BioProductsTM, MBP, catalog number: 7003 ) stored at room temperature
- SYBRTM Safe DNA Gel Stain (Thermo Fisher Scientific, Invitrogen, catalog number: S33102 ) stored at 4 °C
- Complete parasite culture medium (see Recipes)
- Ethanol 75% (see Recipes)
- 10x TBE (see Recipes)
- 2 mM EDTA (see Recipes)
- 1x TBE (see Recipes)
- 1x TBE-SDS 2% (see Recipes)
- 1 kb DNA Ladder (50 µg/ml) (see Recipes)
Equipment
- Autoclaved glass bottles (SCHOTT DURAN)
- -80 °C freezer
- FormaTM Steri-CycleTM CO2 Incubator at 26 °C (Thermo Fisher Scientific, catalog number: 371 )
- Pipette controller (INTEGRA, catalog number: 155017 )
- Pipettes 1,000 µl, 200 µl, 20 µl (Gilson®)
- Pipette 10 µl (Discovery Comfort)
- Laminar flow hood parasite culture (SafeFAST Premium)
- Chemical hood
- Centrifuge 5810 R (Eppendorf) at room temperature and set here at 24 °C
- Centrifuge MICRO STAR 17 (VWR) kept in a cold room at 4 °C
- NanoDropTM ND-1000 Spectrophotometer (ThermoFisher Scientific)
- Water bath (JULABO GmbH)
- Balance (NewClassic MF, MS4002SDR, METTLER TOLEDO)
- Ultra ClearTM (BLANC-LABO) system
Note: The Ultra ClearTM (BLANC-LABO) system delivers Ultra-pure DNase/RNase-free water. - Mini-gel migration system “home-made” (see Figure 2)
- Electrophoresis Power Supply (GIBCO BRL, Life Technologies, ST305) and power cables
- Microwave (FAR)
- Blue LED transilluminator (Dark Reader DR-88X, Clare Chemical Research)
- Camera (Computar iAi CVAA5 IR No C510390B RoHS)
- Video Graphic Printer (SONY, UP-895CE)
Procedure
The steps of the RNA extraction (A) and purification (B) are summarized in Figure 1.
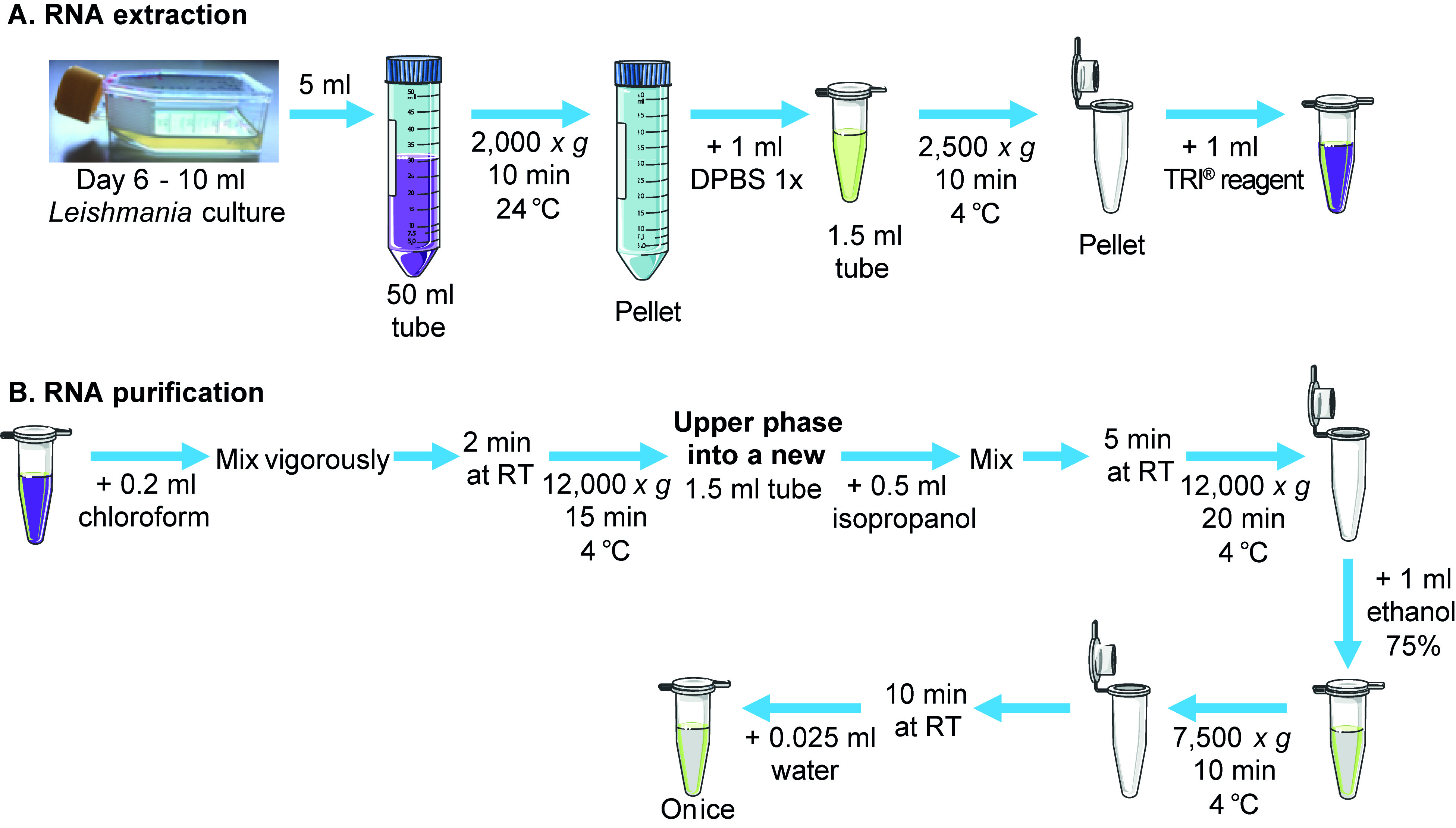
Figure 1. Steps of RNA extraction and purification. This diagram represents the steps of RNA extraction (A) and purification (B). Certain illustrations are from Servier Medical Art, licensed under a Creative Common Attribution 3.0 Unported License. https://smart.servier.com/.
- Total RNA extraction (not DNA-free) from Leishmania parasites (see Figure 1)
Note: The strains are not available commercially. They are derived from Lgy M4147 (MHOM/BR/75/M4147).- Under a laminar flow hood, collect 5 ml of a 10 ml stationary phase parasite culture (day 6 of culture, for a detailed protocol of culture, see pages 3 and 4 of the Bio-protocol by Reverte and Fasel, 2019) in a 50 ml tube.
Note: The authors recommend to use a 50 ml tube even for 5 ml parasites, as discarding the medium from the pellet (at Step A3) is easier with a 50 ml tube. - Centrifuge at 2,000 x g for 10 min at 24 °C.
- Under a laminar flow hood, discard the medium and resuspend the pellet with 1 ml of DPBS 1x.
- Under a laminar flow hood, transfer to a 1.5 ml tube.
- Centrifuge at 2,500 x g for 10 min at 4 °C.
- Under a laminar flow hood, discard the supernatant.
- Resuspend and homogenize the pellet with 1 volume of TRI® reagent (1 ml) under chemical hood, in order to extract RNA from the parasites. At this step, the sample can be kept for a few weeks at -80 °C.
- Under a laminar flow hood, collect 5 ml of a 10 ml stationary phase parasite culture (day 6 of culture, for a detailed protocol of culture, see pages 3 and 4 of the Bio-protocol by Reverte and Fasel, 2019) in a 50 ml tube.
- Total RNA purification (not DNA-free) from Leishmania parasites (see Figure 1)
Note: The following protocol is adapted from the protocol of TRI® reagent provided by the manufacturer.- Clean the bench and the pipettes with RNase AWAYTM reagent.
- If kept at -80 °C, thaw the sample and let equilibrate to room temperature (RT) for 5 min.
- Under chemical hood, add 1/5 volume of chloroform (200 µl).
- Mix vigorously and wait for 2 min at room temperature.
- Centrifuge at 12,000 x g for 15 min at 4 °C.
- Transfer the upper aqueous phase (approximately 500 µl) into a new 1.5 ml tube.
- Add the same volume of isopropanol (here 500 µl) and mix, wait for 5 min at room temperature.
Note: At this step, the purification procedure can be stopped and the tube can be stored at -20 °C overnight. - Centrifuge at 12,000 x g, for 20 min, at 4 °C.
- Discard the supernatant and add 1 volume of Ethanol 75% (1 ml) to the pellet.
- Centrifuge at 7,500 x g, for 10 min, at 4 °C and discard the supernatant.
- Let the pellet dry at room temperature under the chemical hood for approximately 10 min.
Note: Do not over dry the pellet as it may be difficult to dissolve. - Add 25 µl UltraPureTM DNase/RNase-free Distilled Water and vortex briefly to dissolve the pellet at room temperature.
- Put the tube on ice and measure RNA concentration with 1.5 µl RNA with NanoDropTM. Check that the A260/A280 and A260/A230 ratios are of good quality (not contaminated).
Note: Make the blank of the NanoDropTM with 1.5 µl of UltraPureTM DNase/RNase-free Distilled Water. - RNA can be kept at -80 °C for long storage.
Note: Repeated freeze and thaw can decrease the concentration of the RNA and fresh RNA can give better results.
- Enzymatic digestion of total RNA (not DNA-free) from Leishmania parasites: degradation of DNA by DNase I, and single-stranded DNA or RNA by Nuclease S1
- Clean the bench and the pipettes with RNase AWAYTM reagent.
- Warm up the water bath to 37 °C.
- Thaw parasite-isolated total RNA (if frozen) and keep on ice.
Note: If the RNA is frozen, measure RNA concentration with NanoDropTM after it thaws. - In a new 1.5 ml tube, add between 5 and 50 µg of RNA, on ice.
- For 1 reaction, digest the total RNA (not DNA-free) with 60 U of Nuclease S1 in Buffer S1 1x and with 10 U of DNase I in UltraPureTM DNase/RNase-free Distilled Water.
Note: In a new 1.5 ml tube on ice, prepare the enzymatic mix for several reactions: mix DNase I, Nuclease S1, Buffer S1 diluted to 1x and UltraPureTM DNase/RNase-free Distilled Water. Then dispense on RNA. This enzymatic mix has been tested on the range: 5 to 50 µg of RNA. - Prepare an undigested negative control for each sample: in a new 1.5 ml tube, on ice, add between 2 and 5 µg of RNA and dilute in UltraPureTM DNase/RNase-free Distilled Water (final volume of 10 µl).
Note: Different quantities of RNA between digested and undigested samples have been used to avoid huge differences of exposure times between them when acquiring pictures of the gel electrophoresis. - Vortex briefly and spin down.
- Incubate the tubes containing RNA and enzymatic mix, and negative control (without enzymatic mix), in the water bath at 37 °C for 1 h.
Note: During this incubation time, the agarose gel can be prepared. - At the end of the incubation, transfer the tubes with the digested RNA and the undigested negative control on ice.
Note: At this step, the samples can be kept at -80 °C for several weeks.
- Visualization of viral dsRNA by migration on mini-gel electrophoresis
- Rinse the autoclaved glass bottles needed with Ultra-pure DNase/RNase-free water, type Ultra ClearTM.
- Prepare 1x TBE solution (Recipe 5) in an autoclaved glass bottle.
- Prepare 1x TBE containing 2% SDS solution (Recipe 6) in an autoclaved glass bottle.
- Clean the material for mini-gel electrophoresis (tank, spacers and comb) with 1x TBE containing 2% SDS solution, then rinse very well with Ultra-pure DNase/RNase-free water, type Ultra ClearTM and with RNase AWAYTM reagent.
- Prepare 0.8% agarose gel.
- Weigh agarose powder in an autoclaved glass bottle: 0.4 g for 1 mini-gel.
- Add 1x TBE: 50 ml for 1 mini-gel.
- Heat with a microwave until agarose is homogeneously dissolved.
Note: Monitor the aspect of the mixture and be careful of the high temperature. - Cool down at room temperature for a few min.
- Mount the mini-gel electrophoresis (see Figure 2).
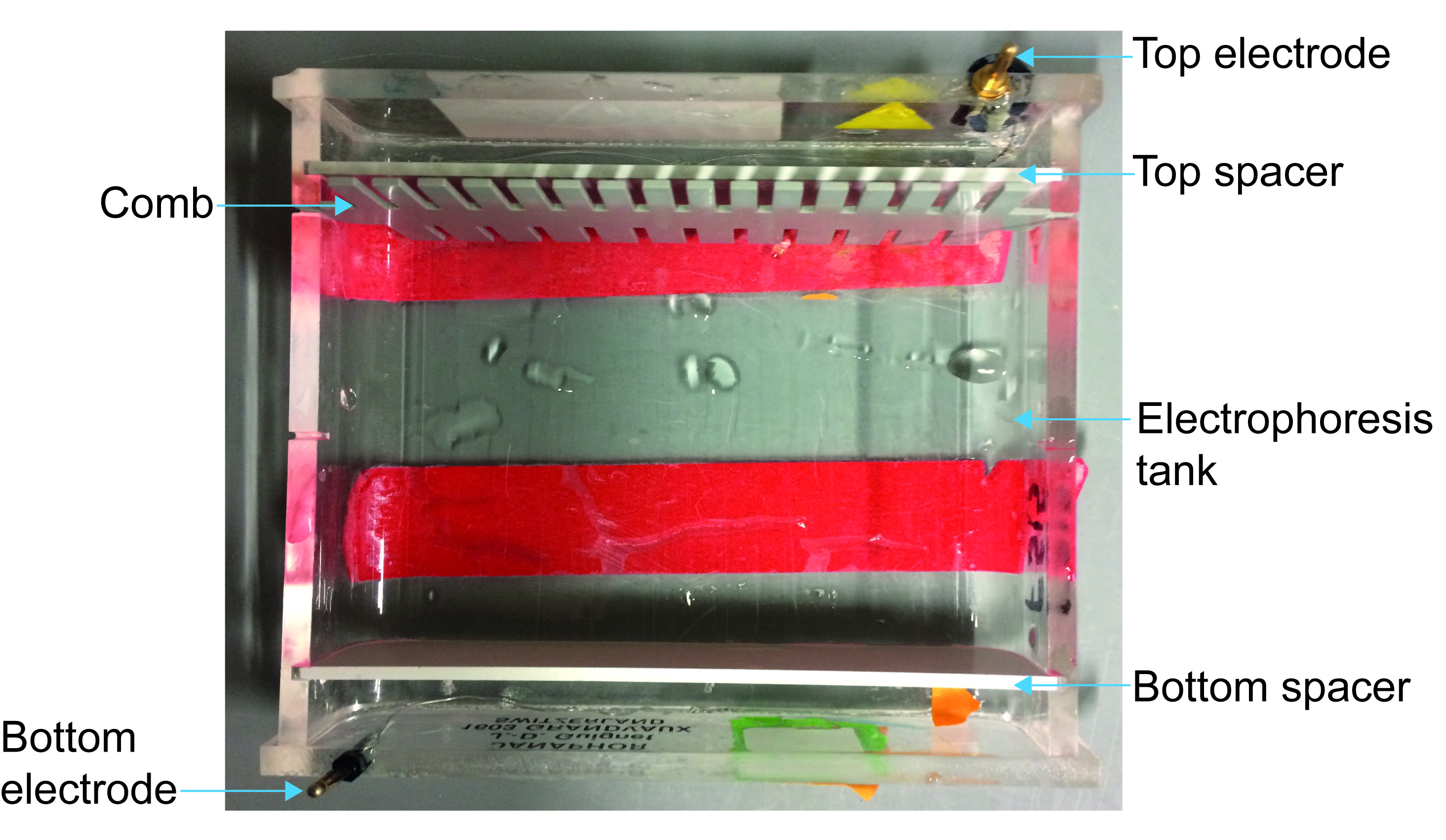
Figure 2. Mini-gel migration system “home-made” for gel electrophoresis. This figure represents the different pieces to assemble before putting the agarose gel. - Add SYBRTM Safe to agarose gel (0.5 µl for 1 mini-gel), shake well but without making bubbles and fill the electrophoresis tank.
- Remove potential bubbles with a tip and let the agarose gel polymerize for 30-45 min at room temperature.
- Pour 1x TBE in the electrophoresis tank (75 ml for 1 mini-gel) and carefully remove the spacers and the comb.
- Add 6x Gel Loading Dye to the samples: the digested RNA and the undigested negative control in order to have Gel Loading Dye at 1x.
- Load the wells with 10-25 µl of each sample and 2-4 µl of 1 kb DNA Ladder (50 µg/ml).
- Connect the electrophoresis tank to the power supply with cables: the top electrode is connected to the negative pole and the bottom electrode to the positive pole.
- Switch on the power supply: run the gel at 80-100 volts for 45-60 min, until the Gel Loading Dye has reached the ¾ of the mini-gel.
- Switch off the power supply and disconnect the electrophoresis tank.
- Transfer the gel to a transilluminator coupled to camera and printer, adjust the zoom, the focus and the exposure time, then take a picture of the mini-gel (see Figure 3).
- Observe and analyse the picture of the gel electrophoresis (see Data analysis).
Data analysis
In the figure of the gel electrophoresis, in the digested RNA (+) lanes (see Figure 3), an upper band at around 5 kb (see blue arrow) is observed in the digested RNA sample from LRV+ parasites, and not in the digested RNA sample from LRV- parasites. The theoretical size of LRV is 5.3 kb. The DNase I degrades dsDNA and ssDNA of the RNA samples that were not free from DNA, whilst the Nuclease S1 degrades single-stranded nucleic acids, such as ssRNA and ssDNA. In Figure 3, the viral dsRNA from LRV (see blue arrow) can be visualized after enzymatic digestions because DNase I and Nuclease S1 do not degrade dsRNA. In the undigested negative controls (-), 3 major bands between 1.5 and 2 kb are observed, they represent the 3 ribosomal RNAs (rRNA), which are ssRNA. The dsRNA at around 5 kb is also observed (see blue arrow). There can also be other bands corresponding to contaminant DNA.
Note: To go further, it is also possible to treat with RNase III enzyme for 20 min at 37 °C. The RNase III degrades dsRNA. Following gel electrophoresis, the band at around 5 kb will no longer be visible.
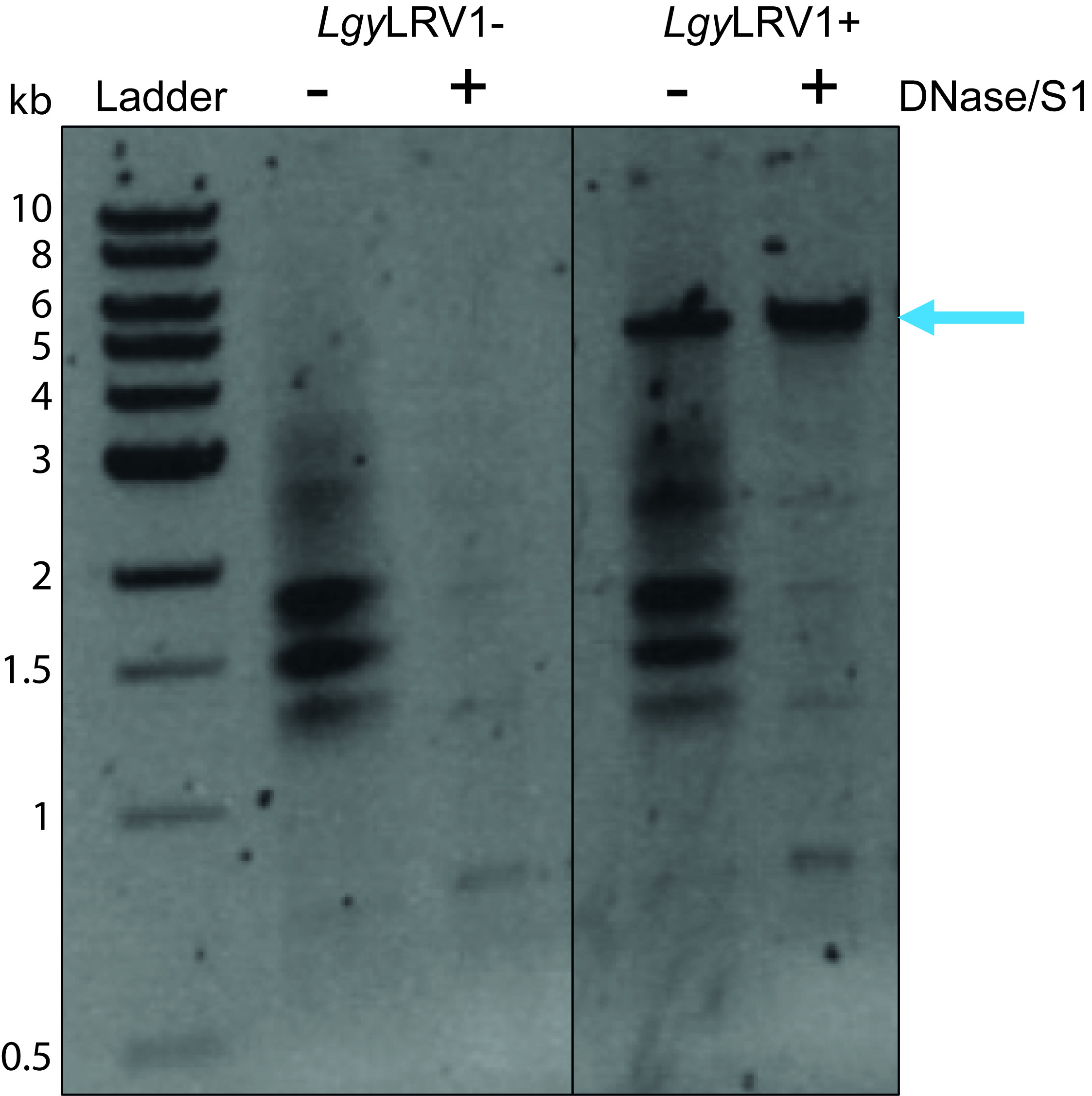
Figure 3. Image of the gel at the end of electrophoresis. The Ladder represents the 1 kb DNA Ladder loaded into the first well on the left. The scale in kilobases (kb) is indicated (from 10 to 0.5 kb). The negative controls without enzymatic digestions are represented by (-). The samples, which are digested, are indicated with (+). The black dividing line (between LgyLRV1- samples and LgyLRV1+ samples) represents where the splice junction has been made. The blue arrow indicates the band of the dsRNA. Lgy: Leishmania guyanensis; LRV1: Leishmania RNA Virus; LRV1-: negative for LRV presence; LRV1+: positive for LRV presence.
Recipes
- Complete parasite culture medium (500 ml)
- Remove 111 ml from the 500 ml Schneider’s Drosophila Medium.
- Then add 100 ml heat-inactivated FBS (see Recipes from Bio-protocol by Reverte and Fasel, 2019), 5 ml HEPES, 5 ml P/S, 250 μl Biopterin (see Recipes from Bio-protocol by Reverte and Fasel, 2019), 1 ml Hemin-folate (see Recipes from Bio-protocol by Reverte and Fasel, 2019).
- Filter the complete parasite culture medium using Vacuum Filtration 500–Filtermax, aliquot to 50 ml tubes and store at 4 °C.
- Ethanol 75%
- For 50 ml, in a 50 ml tube, add 17.5 ml Ultra-pure DNase/RNase-free water, type Ultra ClearTM with a 25 ml pipette connected to a pipette controller.
- Then add 37.5 ml Ethanol absolute.
- Store at 4 °C.
- 10x TBE (1 L)
- Add 108 g Tris(hydroxymethyl)aminomethane, 55 g boric acid and 2 mM EDTA (Recipe 4)
- Add water to 1 L
- Store at room temperature
- 2 mM EDTA (1 L)
- Add 0.74446 g EDTA powder
- Add water to 1 L
- Store at room temperature
- 1x TBE (1 L)
- Add 100 ml 10x TBE (Recipe 3) and 900 ml water
- Store at room temperature
- 1x TBE-SDS 2%
- For 250 ml, add 25 ml SDS 20% and 225 ml 1x TBE (Recipe 5)
- Store at room temperature
- 1 kb DNA Ladder (50 µg/ml)
- For 240 µl, in a 1.5 ml tube, on ice, add 176 µl of UltraPureTM DNase/RNase-free Distilled Water
- Add 40 µl of 6x Gel Loading Dye and add 24 μl of 1 kb DNA Ladder at 500 µg/ml
- Store at 4 °C
Acknowledgments
We thank Slavica Masina and Tiia Snäkä for critical reading of the manuscript; Marta Reverte for helping with several catalog numbers already referenced (see Bio-protocol by Reverte and Fasel, 2019); Chantal Desponds and Florence Prével for providing technical advice on protocol development. This research was supported by FNS grant No 310030_173180 and No IZRJZ3_164176/1 to NF. This protocol was adapted from the following original research papers by Ives et al., 2011; Zangger et al., 2013 and 2014; Grybchuk et al., 2018.
Competing interests
The authors declare no competing interests.
References
- Adaui, V., Lye, F., Akopyants, N. S., Zimic, M., Llanos-Cuentas, A., Garcia, L., Maes, I., De Doncker, S., Dobson, D. E., Arevalo, J., Dujardin, J. C. and Beverley, S. M. (2016). Association of the endobiont double-stranded RNA virus LRV1 with treatment failure for human Leishmaniasis caused by Leishmania braziliensis in Peru and Bolivia. J Infect Dis 213(1): 112-121.
- Akopyants, N. S., Lye, L. F., Dobson, D. E., Lukes, J. and Beverley, S. M. (2016). A novel Bunyavirus-like virus of trypanosomatid protist parasite. Genome Announc 4(4): e00715-16.
- Bourreau, E., Ginouves, M., Prévot, G., Hartley, M. A., Gangneux, J. P., Robert-Gangneux, F., Dufour, J., Sainte-Marie, D., Bertolotti, A., Pratlong, F., Martin, R., Schütz, F., Couppié, P., Fasel, N. and Ronet, C. (2016). Presence of Leishmania RNA Virus 1 in Leishmania guyanensis increases the risk of first-line treatment failure and symptomatic relapse. J Infect Dis 213(1): 105-111.
- El-Gayar, E. K., Mokhtar, A. B. and Hassan, W. A. (2016). Molecular characterization of double-stranded RNA virus in Trichomonas vaginalis Egyptian isolates and its association with pathogenicity. Parasitol Res 115(10): 4027-4036.
- Fernandez-Presas, A. M., Padilla-Noriega, L., Becker, I., Robert, L., Jiménez, J. A., Solano, S., Delgado, J., Tato, P. and Molinari, J. L. (2017). Enveloped and non-enveloped viral-like particles in Trypanosoma cruzi epimastigotes. Rev Inst Med Trop Sao Paulo 59: e46.
- Fichorova, R. N., Buck, O. R., Yamamoto, H. S., Fashemi, T., Dawood, H. Y., Fashemi, B., Hayes, G. R., Beach, D. H., Takagi, Y., Delaney, M. L., Nibert, M. L., Singh, B. N. and Onderdonk, A. B. (2013). The villain team-up or how Trichomonas vaginalis and bacterial vaginosis alter innate immunity in concert. Sex Transm Infect 89(6): 460-466.
- Ghosh, S., Banerjee, P., Sarkar, A., Datta, S. and Chatterjee, M. (2012). Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J Clin Microbiol 50(8): 2774-2778.
- Grybchuk, D., Akopyants, N. S., Kostygov, A. Y., Konovalovas, A., Lye, L. F., Dobson, D. E., Zangger, H., Fasel, N., Butenko, A., Frolov, A. O., Votypka, J., d’Avila-Levy, C. M., Kulich, P., Moravcova, J., Plevka, P., Rogozin, I. B., Serva, S., Lukes, J., Beverley, S. M. and Yurchenko, V. (2018). Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc Natl Acad Sci U S A 115(3): E506-E515.
- Hartley, M. A., Bourreau, E., Rossi, M., Castiglioni, P., Eren, R. O., Prevel, F., Couppié, P., Hickerson, S. M., Launois, P., Beverley, S. M., Ronet, C. and Fasel, N. (2016). Leishmaniavirus-dependent metastatic Leishmaniasis is prevented by blocking IL-17A. PLoS Pathog 12(9): e1005852.
- Ives, A., Ronet, C., Prevel, F., Ruzzante, G., Fuertes-Marraco, S., Schutz, F., Zangger, H., Revaz-Breton, M., Lye, L. F., Hickerson, S. M., Beverley, S. M., Acha-Orbea, H., Launois, P., Fasel, N. and Masina, S. (2011). Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 331(6018): 775-778.
- Lye, L. F., Akopyants, N. S., Dobson, D. E. and Beverley, S. M. (2016). A narnavirus-like element from the trypanosomatid protozoan parasite leptomonas seymouri. Genome Announc 4(4): e00713-16.
- Rath, C. T., Schnellrath, L. C., Damaso, C. R., de Arruda, L. B., Vasconcelos, P. F. D. C., Gomes, C., Laurenti, M. D., Calegari Silva, T. C., Vivarini, A. C., Fasel, N., Pereira, R. M. S. and Lopes, U. G. (2019). Amazonian Phlebovirus (Bunyaviridae) potentiates the infection of Leishmania (Leishmania) amazonensis: Role of the PKR/IFN1/IL-10 axis. PLoS Negl Trop Dis 13(6): e0007500.
- Reverte, M. and Fasel, N. (2019). Leishmania Parasite Quantification by Bioluminescence in Murine Models. Bio-protocol 9(22): e3431.
- Rossi, M., Castiglioni, P., Hartley, M. A., Eren, R. O., Prével, F., Desponds, C., Utzschneider, D. T., Zehn, D., Cusi, M. G., Kuhlmann, F. M., Beverley, S. M., Ronet, C. and Fasel, N. (2017). Type I interferons induced by endogenous or exogenous viral infections promote metastasis and relapse of leishmaniasis. Proc Natl Acad Sci U S A 114(19): 4987-4992.
- Vieira-Gonçalves, R., Fagundes-Silva, G. A., Heringer, J. F., Fantinatti, M., Da-Cruz, A. M., Oliveira-Neto, M. P., Guerra, J. A. O. and Gomes-Silva, A. (2019). First report of treatment failure in a patient with cutaneous leishmaniasis infected by Leishmania (Viannia) naiffi carrying Leishmania RNA virus: a fortuitous combination? Rev Soc Bras Med Trop 52: e20180323.
- Wang, A. L. and Wang, C. C. (1991). Viruses of parasitic protozoa. Parasitol Today 7(4): 76-80.
- Zangger, H., Hailu, A., Desponds, C., Lye, L. F., Akopyants, N. S., Dobson, D. E., Ronet, C., Ghalib, H., Beverley, S. M. and Fasel, N. (2014). Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis 8(4): e2836.
- Zangger, H., Ronet, C., Desponds, C., Kuhlmann, F. M., Robinson, J., Hartley, M. A., Prevel, F., Castiglioni, P., Pratlong, F., Bastien, P., Müller, N., Parmentier, L., Saravia, N. G., Beverley, S. M. and Fasel, N. (2013). Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis 7(1): e2006.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Isorce, N. and Fasel, N. (2020). Viral Double-Stranded RNA Detection by DNase I and Nuclease S1 digestions in Leishmania parasites. Bio-protocol 10(9): e3598. DOI: 10.21769/BioProtoc.3598.
Category
Microbiology > Pathogen detection > PCR
Microbiology > Microbial biochemistry > RNA
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link