- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Auxin-mediated Protein Degradation in Caenorhabditis elegans
Published: Vol 10, Iss 8, Apr 20, 2020 DOI: 10.21769/BioProtoc.3589 Views: 5743
Reviewed by: Gal HaimovichManish ChamoliJian Chen

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Heterochronic Phenotype Analysis of Hypodermal Seam Cells in Caenorhabditis elegans
Yon Ju Ji and Jiou Wang
Jan 5, 2019 6534 Views

Labelling of Active Transcription Sites with Argonaute NRDE-3—Image Active Transcription Sites in vivo in Caenorhabditis elegans
Antoine Barrière and Vincent Bertrand
Jun 5, 2022 2683 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 93 Views
Abstract
The auxin-inducible degron (AID) technology was recently adapted for use in the nematode Caenorhabditis elegans. Rapid degradation of C. elegans proteins tagged with an AID is mediated by a plant-specific F-box protein, transport inhibitor response 1 (TIR1), and occurs only in the presence of the phytohormone auxin. The first iteration of this technology elicited protein degradation in C. elegans through a naturally occurring form of auxin, indole-3-acetic acid (IAA). Here, we present a protocol that uses 1-naphthaleneacetic acid, potassium salt (K-NAA), an indole-free synthetic auxin analog. At equal concentration, K-NAA is as effective as IAA in standard nematode growth media (NGM). K-NAA is also effective in physiological buffer (M9), allowing for high-throughput experimentation. The main advantages of K-NAA are twofold: first, its photostability prevents light-induced compound degradation during storage and the production of toxic indole-derivatives during fluorescence microscopy of live cells; and second, its water solubility eliminates the need of using ethanol to dissolve the auxin compound, a solvent that may confound C. elegans lifespan and behavioral assays. In this protocol, we describe our method of degrading C. elegans proteins using K-NAA on solid and in liquid media, as well as our method of analyzing protein degradation.
Keywords: AID technologyBackground
Conditional protein degradation through degrons is an emerging method for studying protein function in Caenorhabditis elegans (Nance and Frøkjær-Jensen, 2019). Current degron methods include ZF1 (Armenti et al., 2014; Sallee et al., 2018), auxin-inducible degron (AID) (Zhang et al., 2015; Martinez et al., 2019), and GFP nanobody-mediated protein degradation (Wang et al., 2017). Originally, the auxin-based degron method was co-opted from plants and transplanted into nonplant systems such as yeast, chicken and mammalian cell culture (Nishimura et al., 2009). This method was recently optimized for use in C. elegans due to advances in CRISPR/Cas9 gene-editing technology (Zhang et al., 2015; Dickinson and Goldstein, 2016). In the presence of the plant hormone auxin, C. elegans proteins tagged with an AID are rapidly degraded by a heterologously expressed plant-specific F-box protein, transport inhibitor response 1 (TIR1). TIR1 functions as a substrate-recognition component of the CUL-1-based SCF E3 ubiquitin ligase complex consisting of SKR-1/2, CUL-1 and the RING component RBX-1 (Martinez et al., 2019). In as little as 30 min, C. elegans proteins exposed to auxin can be precisely degraded in a tissue- and cell-specific manner (Zhang et al., 2015; Martinez et al., 2019).
Either the natural auxin indole-3-acetic acid (IAA) or the synthetic auxin 1-naphthaleneacetic acid (NAA) can be used to conditionally deplete target proteins (Zhang et al., 2015; Martinez et al., 2019). However, NAA has the advantage over IAA partly due to its photostability (Yamakawa et al., 1979). Blue light, used to excite GFP, in combination with IAA has been shown to cause defects in mitosis and meiosis in yeast and mammalian oocytes, respectively (Papagiannakis et al., 2017; Camlin and Evans, 2019). Such effects likely occur through the photo-destruction of IAA to its toxic indole derivatives (Folkes and Wardman, 2001; Srivastava, 2002). Furthermore, the potassium salt of NAA (K-NAA) has the extra advantage of being completely water-soluble. This circumvents the need to expose C. elegans to low percentages of ethanol, used to dissolve IAA, or other potentially harmful solvents (Li et al., 2019). Additionally, when compared to 4 mM IAA, an equivalent concentration of K-NAA does not affect growth of C. elegans bacterial food Escherichia coli OP50 (Martinez et al., 2019). Also, 4 mM IAA results in significant embryonic lethality compared to K-NAA of the same concentration (Martinez et al., 2019). Potential applications of this protocol include high-throughput, high-resolution chemical genetic screens using a microfluidic device or a 96-well plate where worms are immersed in K-NAA. The protocol described here will elaborate on a simple procedure where C. elegans larvae are immersed in K-NAA diluted in M9 physiological buffer on a spot plate or exposed to K-NAA diluted in traditional nematode growth media (NGM). Finally, a step-by-step procedure to quantify the extent of protein degradation is presented.
Materials and Reagents
- Laboratory labeling tape
- 60 x 15 mm medium size Petri dish (Crystalgen, catalog number: S-3004 )
- 90% platinum, 10% iridium wire (Tritech Research, catalog number: PT-9010 )
- 0.1-10 μl micropipette tips (VWR, catalog number: 46620-316 )
- 1-200 μl micropipette tips (VWR, catalog number: 53508-783 )
- 100-1,250 μl micropipette tips (VWR, catalog number: 53508-918 )
- 15 ml centrifuge tubes (Thermo Fisher Scientific, catalog number: 339650 )
- 50 ml centrifuge tubes (VWR, catalog number: 89401-572 )
- 30 ml syringe (Fisher Scientific, catalog number: 309650 )
- Sterile syringe filter, 5 μm, 25 mm (Pall, catalog number: 4199 )
- Spot plate (Thomas Scientific, catalog number: 7812G17 )
- 9 ml glass culture tubes (VWR, catalog number, 47729-572 )
- 2 ml glass pipette (VWR, catalog number: 14673-043 )
- 18 x 18 mm, #1.5 thickness microscope cover slip (Fisherbrand, catalog number: 12-541A )
- 25 x 75 x 1.0 mm microscope slides (Fisherbrand, catalog number: 12-550-A3 )
- Paper towels
- 1.5 ml microcentrifuge tubes
- Worm pick handle (Tritech Research, catalog number: TWPH1 )
- Escherichia coli OP50 (University of Minnesota, Caenorhabditis Genetic Center)
- Caenorhabditis elegans CA1202 (control strain for auxin-inducible degradation experiments in somatic tissues) (University of Minnesota, Caenorhabditis Genetic Center, genotype: ieSi57[eft-3>TIR1::mRuby] II; ieSi58[eft-3>AID::GFP] IV)
- 6.0% sodium hypochlorite solution (Ace Hardware, catalog number: 1912120 )
- Naphthaleneacetic acid (K-NAA) (PhytoTechnology Laboratories, catalog number: N610 )
- Mounting agar (VWR, catalog number: 0 815 )
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S2002 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-3 )
- Peptone (Thermo Fisher Scientific, catalog number: 211677 )
- Bacteriological agar (Lab Scientific, catalog number: A466 )
- 100% ethanol (Pharmco, catalog number: 111000200 )
- Double-distilled water (ddH2O)
- Potassium hydroxide (KOH) (BioExpress, catalog number: 0489)
- Cholesterol (BioExpress, catalog number: 0433 )
- Calcium chloride (CaCl2) (Fisher Scientific, catalog number: C79-500 )
- Magnesium sulfate (MgSO4) (Fisher Scientific, catalog number: M63-500 )
- Monopotassium phosphate (KH2PO4) (Fisher Scientific, catalog number: BP362 )
- Dipotassium phosphate (K2HPO4) (Fisher Scientific, catalog number: BP363 )
- Disodium phosphate (Na2HPO4) (Fisher Scientific, catalog number: S374 )
- 5 M KOH stock solution (see Recipes)
- 5 mg/ml cholesterol (see Recipes)
- 1 M CaCl2 stock solution (see Recipes)
- 1 M MgSO4 stock solution (see Recipes)
- 1 M KPO4, pH 6.0 stock solution (see Recipes)
- 250 mM K-NAA stock solution (see Recipes)
- Nematode growth media (NGM) plates (see Recipes)
- NGM plates containing K-NAA (see Recipes)
- 1x M9 buffer (see Recipes)
- K-NAA in 1x M9 buffer (see Recipes)
- 5% agarose (see Recipes)
- 1 M NaN3 stock solution (see Recipes)
- Agarose containing NaN3 (see Recipes)
- Agarose pad containing NaN3 (see Recipes)
Equipment
- 0.1-2 μl micropipette (Thomas Scientific, catalog number: P3960-2A-B )
- 2-20 μl micropipette (Thomas Scientific, catalog number: P3960-20A-B )
- 20-200 μl micropipette (Thomas Scientific, catalog number: P3960-200A-B )
- 100-1,000 μl micropipette (Thomas Scientific, catalog number: P3960-1000A-B )
- Rubber pipette bulb (Thomas Scientific, catalog number: 1951F25 )
- Microwave
- Bunsen burner
- Scale
- 250 ml storage bottles
- 500 ml storage bottles
- 1 L storage bottles
- 4 L Erlenmeyer flask
- Magnetic stirring bar
- Autoclave
- Stir plate (Corning, model: PC-310 )
- Vortex mixer (VWR, catalog number: 10153-838 )
- Microcentrifuge (Eppendorf, model: Centrifuge 5424 )
- Benchtop centrifuge (Beckman Coulter, model: Allegra X-12R )
- Tube rocker (Benchmark Scientific, model: M2100 )
- Dry bath incubator (Fisher Scientific, catalog number: 11-718 )
- Dissecting microscope (Zeiss, model: Stemi 508 )
- EM-CCD camera (Hamamatsu, model: C9100-23B )
- Spinning disk confocal microscope (Yokogawa, model: CSU10 )
Software
- ImageJ (version: 2.0.0-rc-69/1.52p)
Procedure
- L1 stage synchronization by sodium hypochlorite treatment
Note: Synchronization via egg lay is an alternative method that can be used (Gidalevitz et al., 2009).- Prepare NGM plates (Recipe 7).
- Seed NGM plates with 200-300 μl of overnight grown E. coli OP50 under sterile conditions. Allow the plates to dry at room temperature for approximately 48 h. Store at 4 °C until use. Warm plates at room temperature before use.
- Using a worm picker, pick the desired number of L4 stage CA1202 worms, or any AID-tagged strain of interest, onto NGM plates seeded with E. coli OP50. Allow the worms to grow until adulthood. Depending on the rate of usage, grow between 15-25 °C (Stiernagle, 2006).
- Recover gravid adults in a 15 ml centrifuge tube by washing plates with ddH2O.
- Add ddH2O to a total volume of 8 ml.
- Add 600 μl of 5 M KOH.
- Add 1,200 μl of 6.0% sodium hypochlorite solution.
- Place worms on a low speed tube rocker for 7-9 min.
- Pellet released eggs by centrifuging for 1 min at 400 x g at room temperature using a benchtop centrifuge. Quickly but carefully decant the supernatant.
Note: Use a balancer containing 9.8 ml of ddH2O, if needed. - Add 10 ml of ddH2O to the pellet. Gently mix the tube by inverting 3-4 times.
- Pellet released eggs by centrifuging for 2 min at 400 x g at room temperature using a benchtop centrifuge. Quickly but carefully decant the supernatant.
Note: Use a balancer containing 10 ml of ddH2O, if needed. - Add 10 ml of 1x M9 buffer to the pellet. Gently mix the tube by inverting 3-4 times.
- Repeat Step A11.
- Add 1x M9 buffer to the pellet to a final volume of 3 ml and incubate eggs at 20 °C on a low speed tube rocker.
- After an incubation time of 16 h, add the desired concentration of L1 worms to 2-3 NGM plates seeded with E. coli OP50.
- Allow worms to grow to the desired developmental stage using standard growth conditions (Stiernagle, 2006).
- Auxin treatment on solid media
- Prepare NGM plates containing 1 mM or 4 mM K-NAA (Recipe 8).
- Seed NGM plates containing K-NAA with 200-300 μl of overnight grown E. coli OP50 under sterile conditions. Allow the plates to dry at room temperature for approximately 48 h. Store at 4 °C until use. Warm plates at room temperature before use.
- Once at the desired developmental stage, recover synchronized worms in 1.5 ml microcentrifuge tubes by washing plates with 500 μl of 1x M9 buffer.
- Gently pellet worms by centrifuging for 2 min at 200 x g at room temperature using a microcentrifuge.
- Carefully remove as much of the supernatant as possible with a micropipette and discard it.
- Mix the remaining contents (i.e., worms and residual bacterial food) by gently tapping the bottom of the tube.
- Evenly dispense the contents onto 2-3 NGM plates containing K-NAA seeded with E. coli OP50 (100-200 worms per plate).
- Wait as little as 30 min to observe auxin-induced degradation of AID-tagged C. elegans proteins under a spinning disk confocal microscope (Figure 1). At this time, review the preparation of an agarose pad containing NaN3 (Recipe 14 and Procedure D).
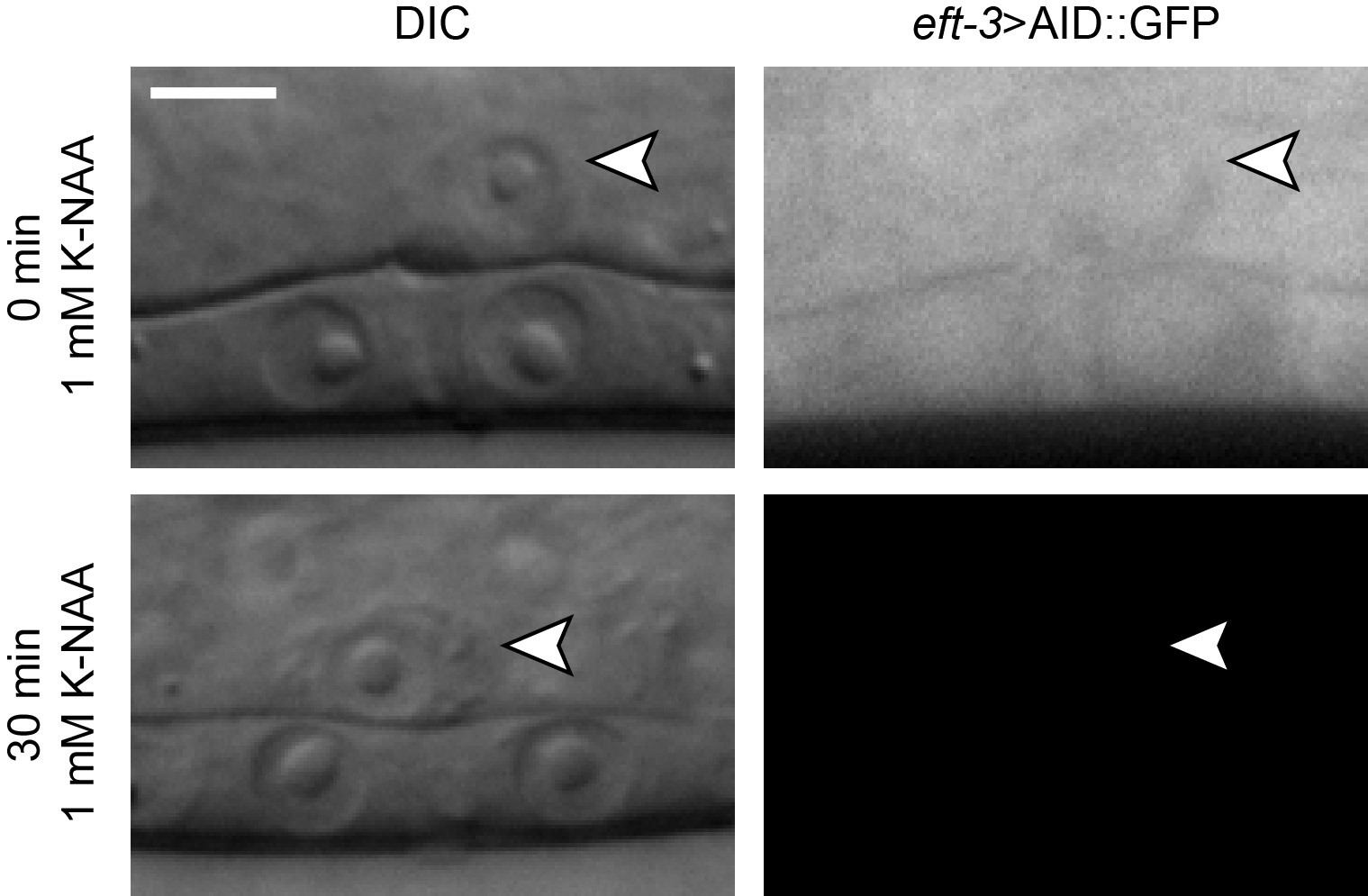
Figure 1. K-NAA-mediated degradation in C. elegans. Differential interference contrast (DIC) and corresponding green fluorescent protein (GFP) images of uterine anchor cells (ACs) (arrowheads) at the mid-L3 stage of C. elegans larval development. Scale bar, 5 μm.
- Auxin treatment in liquid media
- Prepare 1 mM or 4 mM K-NAA in 1x M9 buffer (Recipe 10). Store at 4 °C until use. Warm the solution at room temperature before use.
- Prepare an in-house humidity incubation box (Figure 2).

Figure 2. An in-house humidity incubation box. Wet paper towels are placed along the four bottom edges of an empty pipette box and dry paper towel is attached to the inner surface of the lid. The wet paper towels should create a pocket for a spot plate to fit in. - Once at the desired developmental stage, recover synchronized worms (100-200 worms per well) on a spot plate by washing plates with 500 μl of K-NAA in 1x M9 buffer.
Note: Directly washing worms off plates transfers the bacterial food source, preventing short-term starvation (~2 h). Additional bacterial food can be re-suspended as needed. We recommend allowing the worms to thrash freely on a spot plate and not in a 1.5 ml microcentrifuge tube as it decreases degradation efficiency in our hands. - Place the spot plate in the in-house humidity incubation box (Figure 2). Close the lid on the box.
- Wait as little as 30 min to observe auxin-induced degradation of AID-tagged C. elegans proteins under a spinning disk confocal microscope (Figure 1). At this time, review the preparation of an agarose pad containing NaN3 (Recipe 14 and Procedure D).
- Worm mounting and anesthetization
- After 30 min, gently break apart the two microscope slides interfacing with the agarose pad containing NaN3. Use the microscope slide with the agarose pad containing anesthetic.
Note: We recommend using the agarose pad within 5 min to prevent desiccation. - For auxin treatment on solid media, add 1.2 μl of 1x M9 buffer to the center of the pad. Pick 30-50 worms and gently dab them on the M9. Use a dissecting microscope to ensure the worms have fully transferred off the worm picker.
- For auxin treatment in liquid media, remove 1.2-2 μl with a micropipette from the center of the spot plate well and drop the worms onto the center of the pad.
- Gently place a microscope cover slip over the agar pad.
Note: To avoid air bubbles underneath the cover slip, place the cover slip at a 45-degree angle and gently drop the cover slip over the agar pad. The cover slip should be applied gently to avoid vulval expulsion. We do not recommend pressing down on the cover slip once it has been released.
- After 30 min, gently break apart the two microscope slides interfacing with the agarose pad containing NaN3. Use the microscope slide with the agarose pad containing anesthetic.
- Worm imaging
- Place the microscope slide on the stage and secure it with the stage clips.
- Move the 10x objective lens into position.
- Focus on the worm using the coarse focus knob.
- Finish focusing on the worm using the fine focus knob.
- Place a small drop of oil on the cover slip.
- Move the 100x objective lens into position.
- Slowly focus on the tissue or cell of interest using the fine focus knob.
- Collect 16-bit z-stack images using an EM-CCD camera attached to a spinning disk confocal microscope.
- Imaging parameters (e.g., exposure settings and laser power) should be documented and kept consistent.
Data analysis
- Open ImageJ software (version: 2.0.0-rc-69/1.52p).
- Click Analyze > Set Measurements. Make sure that mean gray value is selected.
- Drag and drop TIFF file containing DIC z-stack image.
- Scroll through DIC z-stack image to find the cell of interest.
- Zoom in on the cell of interest.
- Use the freehand selection tool to outline the cell of interest (Figure 3).
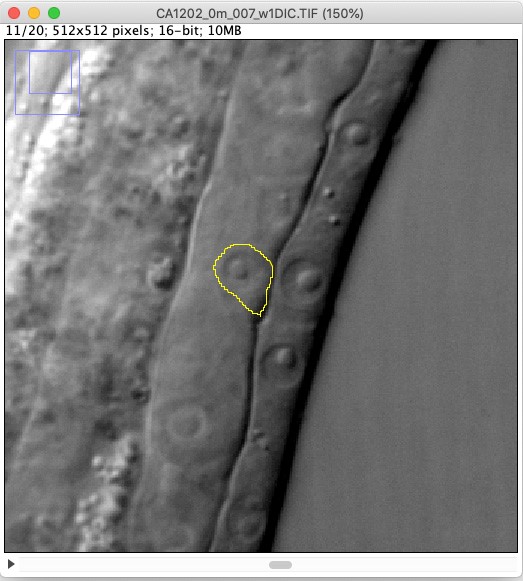
Figure 3. A screenshot image of an outlined AC revealed by DIC microscopy - Click Analyze > Tools > ROI Manager.
- Click Add[t].
- Drag and drop TIFF file containing corresponding fluorescent protein z-stack image.
- Click the ROI (Figure 4).
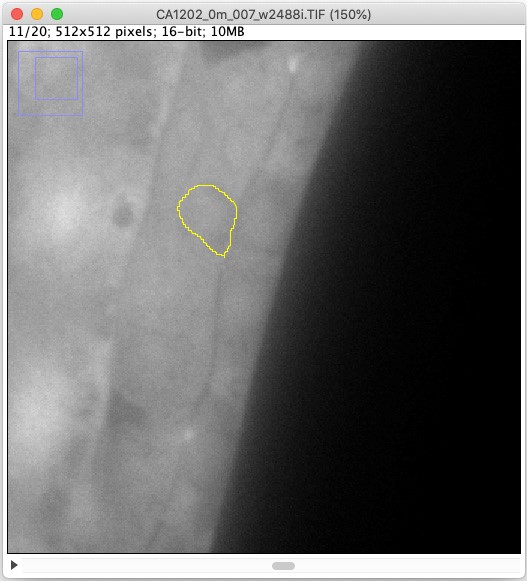
Figure 4. A screenshot image of an outlined AC in a transgenic worm expressing eft-3>AID::GFP
- Click Analyze > Measure.
- Drag freehand selection to a background region in the image.
- Click Analyze > Measure.
- Subtract the mean gray value of the background from the mean gray value of the cell of interest. This subtraction accounts for camera noise.
- Document the calculated value in a spreadsheet and repeat analysis for each imaged worm.
- Normalize the data by dividing the cell’s mean gray value upon auxin treatment by the cell’s mean gray value prior to auxin treatment.
- Perform a minimum of three independent experiments per treatment condition examining at least 20-30 worms per experiment.
Notes
It has not been tested whether a higher concentration of K-NAA (> 4 mM) will inhibit E. coli OP50 growth, or whether it will lead to faster depletion of AID-tagged proteins in C. elegans (Martinez et al., 2019). By and large, degradation rates are similar in the presence of 1 mM and 4 mM auxin (Zhang et al., 2015; Martinez et al., 2019). Some endogenously tagged proteins seem to respond better to 4 mM auxin, such as the nuclear hormone receptor NHR-25 (Martinez et al., 2019). However, we currently believe that the rate-limiting factor in the AID system is the auxin receptor TIR1.
Recipes
- 5 M KOH stock solution
- Dissolve 280.55 g of KOH in 1 L of ddH2O
- Aliquot 200 ml into 500 ml storage bottles and store at room temperature
- Dissolve 280.55 g of KOH in 1 L of ddH2O
- 5 mg/ml cholesterol
- Dissolve 2.5 g of cholesterol in 500 ml of 100% ethanol
- Mix at 30 °C for 15-30 min to dissolve
- Aliquot 100 ml into 250 ml storage bottles
- Store at room temperature
- Dissolve 2.5 g of cholesterol in 500 ml of 100% ethanol
- 1 M CaCl2 stock solution
Dissolve 73.5 g of CaCl2 in 500 ml of ddH2O and autoclave for 30 min
Store at room temperature - 1 M MgSO4 stock solution
Dissolve 60.2 g of MgSO4 in 500 ml of ddH2O and autoclave for 30 min
Store at room temperature - 1 M KPO4, pH 6.0 stock solution
- Dissolve 68.0 g of KH2PO4 in 500 ml of ddH2O
- Dissolve 52.3 g of K2HPO4 in 300 ml of ddH2O
- While measuring the pH of 1 M KH2PO4, add and stir 1 M K2HPO4 slowly until the pH rises from 4.0 to 6.0
- Aliquot and autoclave for 30 min
- Store at room temperature
- Dissolve 68.0 g of KH2PO4 in 500 ml of ddH2O
- 250 mM K-NAA stock solution
- Dissolve 2.243 g of K-NAA in 40 ml of ddH2O in a 50 ml centrifuge tube and vortex at room temperature to fully dissolve the compound
- Clarify solution through a sterile syringe filter connected to a 30 ml syringe into a new 50 ml centrifuge tube
- Store at 4 °C for up to six months
- Dissolve 2.243 g of K-NAA in 40 ml of ddH2O in a 50 ml centrifuge tube and vortex at room temperature to fully dissolve the compound
- NGM plates
- Add 6 g of NaCl, 5 g of peptone, 34 g of bacteriological agar, and a magnetic stirring bar to a 4 L Erlenmeyer flask
- Add 1,944 ml of ddH2O
- Autoclave for 30 min
- Let the NGM cool to 55-60 °C.
- Add 2 ml of 5 mg/ml cholesterol, 2 ml of 1 M CaCl2, 2 ml of 1 M MgSO4, and 50 ml of 1 M KPO4, pH 6.0
- Thoroughly mix the medium in the flask on a stir plate
- Dispense 8 ml per 60 x 15 mm Petri dish and allow them to solidify for 48 h
- Add 6 g of NaCl, 5 g of peptone, 34 g of bacteriological agar, and a magnetic stirring bar to a 4 L Erlenmeyer flask
- NGM plates containing K-NAA
- Add K-NAA to a desired final concentration immediately prior to dispensing the autoclaved NGM
- For 1 mM K-NAA, add 32 μl of 250 mM K-NAA. For 4 mM K-NAA, add 128 μl of 250 mM K-NAA
- Plates can be stored at 4 °C for up to a month
- Add K-NAA to a desired final concentration immediately prior to dispensing the autoclaved NGM
- 1x M9 buffer
- Add 6 g of Na2HPO4, 3 g of KH2PO4, and 5 g of NaCl to a 1 L storage bottle
- Add 1 L of ddH2O
- Aliquot 200 ml into 500 ml storage bottles and autoclave for 30 min
- Add 200 μl of 1 M MgSO4 to each bottle
- Store at room temperature
- Add 6 g of Na2HPO4, 3 g of KH2PO4, and 5 g of NaCl to a 1 L storage bottle
- K-NAA in 1x M9 buffer
- Dilute K-NAA in 1x M9 buffer to a desired final concentration and final volume of 10 ml
- For 1 mM K-NAA, dilute 40 μl of 250 mM K-NAA in 960 μl of 1x M9 buffer. For 4 mM K-NAA, dilute 160 μl of 250 mM K-NAA in 840 μl of 1x M9 buffer
- Solution can be stored at 4 °C for up to a month
- Dilute K-NAA in 1x M9 buffer to a desired final concentration and final volume of 10 ml
- 5% agarose
- Add 5 g of melting agarose and a magnetic stirring bar to a 250 ml storage bottle
- Add 100 ml of ddH2O
- Microwave until agarose is completely dissolved
- Keep the agarose on a stir plate and aliquot 5 ml into 9 ml glass culture tubes
- Allow it to solidify
- Store at room temperature
- Add 5 g of melting agarose and a magnetic stirring bar to a 250 ml storage bottle
- 1 M NaN3 stock solution
- Dissolve 0.65 g of NaN3 in 10 ml of ddH2O
- Sterilize solution and aliquot into 1.5 ml microcentrifuge tubes
- Store at room temperature for up to 2-3 months
- Dissolve 0.65 g of NaN3 in 10 ml of ddH2O
- Agarose containing NaN3
- Gently microwave a 5 ml aliquot of 5% agarose for 7 s
- Using a Bunsen burner, carefully and thoroughly melt the agarose
- Keep it on a dry bath incubator at 65 °C to prevent it from solidifying
- Add 35 μl of 1 M NaN3 to 5 ml of melted 5% agarose for a final concentration of 7 mM
- Resuspend to mix
- Prepare fresh every 24 h or prior to each imaging session
- Gently microwave a 5 ml aliquot of 5% agarose for 7 s
- Agarose pad containing NaN3
- Carefully snap off the tip from a 2 ml glass pipette
- Fit a rubber pipette bulb on the top of the glass pipette
- Using the rubber pipette bulb attached to a glass pipette, aspirate and drop a small volume of agarose containing NaN3 onto a microscope slide. Avoid bubbles
- Quickly and perpendicularly place a second microscope slide over the slide containing the drop of agarose with anesthetic. Allow the agarose to solidify
Note: We recommend vertically flanking the base microscope slide with taped microscope slides of the same dimension in parallel on a flat level surface. Prepare fresh every 20 min.
- Carefully snap off the tip from a 2 ml glass pipette
Acknowledgments
This protocol was adapted from Martinez et al. (2019). We thank K. Ames for advice on building an in-house humidity incubation box. We also thank the Matus lab, collaborators and colleagues for insightful discussions while developing parts of this protocol. M. Martinez is funded by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) [3R01GM121597-03S1]. D. Matus is also funded by the NIH NIGMS [1R01GM121597-01]. D. Matus receives additional support from the Damon Runyon Cancer Research Foundation [DRR-47-17].
Competing interests
The authors declare no competing interests.
References
- Armenti, S. T., Lohmer, L. L., Sherwood, D. R. and Nance, J. (2014). Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development 141(23): 4640-4647.
- Camlin, N. J. and Evans, J. P. (2019). Auxin-inducible protein degradation as a novel approach for protein depletion and reverse genetic discoveries in mammalian oocytesdagger. Biol Reprod 101(4): 704-718.
- Dickinson, D. J. and Goldstein, B. (2016). CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202(3): 885-901.
- Folkes, L. K. and Wardman, P. (2001). Oxidative activation of indole-3-acetic acids to cytotoxic species- a potential new role for plant auxins in cancer therapy. Biochem Pharmacol 61(2): 129-136.
- Gidalevitz, T., Krupinski, T., Garcia, S. and Morimoto, R. I. (2009). Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet 5(3): e1000399.
- Li, R., Zhan, S., Chen, G., Jin, Y., Yu, B., Zhao, J., Han, D. and Fan, H. (2019). Equilibrium solubility, model correlation, and solvent effect of indole-3-acetic acid in twelve pure solvents. J Chem Eng Data 64(4): 1802-1808.
- Martinez, M. A. Q., Kinney, B. A., Medwig-Kinney, T. N., Ashley, G., Ragle, J. M., Johnson, L., Aguilera, J., Hammell, C. M., Ward, J. D. and Matus, D. Q. (2019). Rapid degradation of Caenorhabditis elegans proteins at single-cell resolution with a synthetic auxin. G3 (Genes, Genomes, Genet) 10(1): 267-280.
- Nance, J. and Frøkjær-Jensen, C. (2019). The Caenorhabditis elegans transgenic toolbox. Genetics 212(4): 959-990.
- Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. and Kanemaki, M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6(12): 917-922.
- Papagiannakis, A., de Jonge, J. J., Zhang, Z. and Heinemann, M. (2017). Quantitative characterization of the auxin-inducible degron: a guide for dynamic protein depletion in single yeast cells. Sci Rep 7(1): 4704.
- Sallee, M. D., Zonka, J. C., Skokan, T. D., Raftrey, B. C. and Feldman, J. L. (2018). Tissue-specific degradation of essential centrosome components reveals distinct microtubule populations at microtubule organizing centers. PLoS Biol 16(8): e2005189.
- Srivastava, L. M. (2002). Chapter 6: Auxins. In: Srivastava L. (Ed.). In: Plant growth and development. 1st Edition. Academic Press, 21-22.
- Stiernagle, T. (2006). Maintenance of C. elegans. WormBook: 1-11.
- Wang, S., Tang, N. H., Lara-Gonzalez, P., Zhao, Z., Cheerambathur, D. K., Prevo, B., Chisholm, A. D., Desai, A. and Oegema, K. (2017). A toolkit for GFP-mediated tissue-specific protein degradation in C. elegans. Development 144(14): 2694-2701.
- Yamakawa, T., Kurahashi, O., Ishida, K., Kato, S., Kodama, T. and Minoda, Y. (1979). Stability of indole-3-acetic acid to autoclaving, aeration and light Illumination. Agric Biol Chem 43(4): 879-880.
- Zhang, L., Ward, J. D., Cheng, Z. and Dernburg, A. F. (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142(24): 4374-4384.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Martinez, M. A. Q. and Matus, D. Q. (2020). Auxin-mediated Protein Degradation in Caenorhabditis elegans. Bio-protocol 10(8): e3589. DOI: 10.21769/BioProtoc.3589.
Category
Developmental Biology > Morphogenesis > Organogenesis
Developmental Biology > Cell growth and fate > Differentiation
Molecular Biology > Protein > Targeted degradation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









