- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vivo Tumor Growth and Spontaneous Metastasis Assays Using A549 Lung Cancer Cells
Published: Vol 10, Iss 7, Apr 5, 2020 DOI: 10.21769/BioProtoc.3579 Views: 8863
Reviewed by: Julie Sandra Di MartinoAna Santos AlmeidaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Immunostaining Protocol: P-Smad2 (Xenograft and Mice)
Alexandre Calon [...] Eduard Batlle
May 5, 2014 9027 Views

Protocol for Murine/Mouse Platelets Isolation and Their Reintroduction in vivo
Jae Hong Im and Ruth J. Muschel
Feb 20, 2017 20491 Views

Generation and Maintenance of Patient-Derived Endometrial Cancer Organoids
Mali Barbi [...] Semir Beyaz
Oct 20, 2024 2094 Views
Abstract
Metastasis accounts for the majority of cancer related deaths. The genetically engineered mouse (GEM) models and cell line-based subcutaneous and orthotopic mouse xenografts have been developed to study the metastatic process. By using lung cancer cell line A549 as an example, we present a modified protocol to establish the cell line-based xenograft. Our protocol ensures sufficient establishment of the mouse xenografts and allows us to monitor tumor growth and spontaneous metastasis. This protocol could be adapted to other types of established cancer cell lines or primary cancer cells to study the mechanism of metastatic process as well as to test the effect of the potential anti-cancer agents on tumor growth and metastatic capacity.
Keywords: Cell line xenograftBackground
The 5-year survival rate for patients with advanced stage lung cancer is less than 15%, with the majority of patients die from metastatic diseases (Heist and Engelman, 2012; Whitsett, et al., 2013; D'Antonio et al., 2014; Steeg, 2016). Therefore, using mouse models to investigate the mechanisms of lung cancer invasion and metastasis could facilitate the development of novel strategy to control the metastatic process.
Metastasis is a multi-step process that includes local invasion, intravasation, and survival in the circulation, extravasation, and establishment of macrometastasis in the distant site. Both mouse xenografts and the genetically engineered mouse (GEM) models have been used to study lung cancer metastasis. The GEM models, mimicking the genetic signature of human lung cancer such as TP53, K-ras and LKB1 provide the immunocompetent system to study the mechanism of metastatic process and to investigate how novel anti-cancer agents affect the process (Dutt and Wong, 2006; Zheng et al., 2007). Patient-derived xenografts (PDX) models take tumor heterogeneity and the contribution of tumor microenvironment into the consideration and maintain the features of the primary patient tumor such as gene expression profiles and drug response (Morton and Houghton, 2007, Okada et al., 2019). Cell line-based xenografts either by subcutaneous or orthotopic injection or implantation of cells into mice are commonly used to study lung cancer metastasis. The orthotopic transplantation of tumor cells into mouse lung resembles most closely to human cancer in the aspect of tumor histology, tumor vasculature, gene expression, and responsiveness to drug treatment as well as the metastatic process (Khanna and Hunter, 2005) and this methodology has been reported previously (Justilien and Fields, 2013). Due to the complexity of the metastatic process, there is no single system that can model all of the steps of metastasis (Khanna and Hunter, 2005). The strength and weakness of different mouse models of metastasis were recently comprehensively reviewed (Gomez-Cuadrado et al., 2017). In this protocol, we used lung cancer cell line A549 as an example and utilized the methodology for generating PDX models to establish cell line-based xenograft and monitor tumor growth and spontaneous metastasis. Unlike the classical subcutaneous injection, we performed surgery on mouse to generate a wide subcutaneous pouch to increase the contact area of the cells and matrigel mixture with the tissue. In addition, the surgical incision is big enough to allow us to use the wide bore pipet tip to implant the cells and matrigel mixture smoothly and to prevent the destruction of the cells. This protocol ensures sufficient establishment of the mouse xenografts subcutaneously and allows us to monitor the formation of micrometastasis. This protocol can be adapted to study the mechanisms of metastatic process using the primary or established cell lines from other cancer types and to test the effect of the potential anti-cancer agents on tumor growth and metastatic capacity.
Materials and Reagents
- 10 cm cell culture dishes (Corning, catalog number: 430167 )
- 1.5 ml Eppendorf tubes
- Pipet tips
- Mouse (NOD.CB17-Prkdcscid/J, The Jackson Laboratory, catalog number: 00 1303 )
- A549 cells (ATCC, catalog number: CCL-185 )
- DMEM/F12 medium (Sigma, catalog number: D8437 )
- Pen/Strep/Glutamine (Gibco, catalog number: 10378-016 )
- Fetal Bovine Serum (FBS) (Sigma, catalog number: F0926 )
- Phosphate-buffered saline (PBS) (Thermo Fisher, catalog number: 10010-049 )
- Growth Factor Reduced Matrigel (Corning, catalog number: 354230 )
- Analgesic (Buprenorphine HCl, 0.1 mg/kg SQ, ZooPharm)
- Trypsin-EDTA (Gibco, catalog number: 25300-054 )
- 70% ethanol (Decon Labs, catalog number: DSP-MD-43)
- 10% neutral buffered formalin (Sigma, catalog number: HT501128 )
- Hematoxylin (VWR, catalog number: 97062-216)
- Eosin (VWR, catalog number: 97061-034)
Equipment
- Sorvall Legend X1 Centrifuge(Thermo Scientific, catalog number: 75004221)
- Vi-cell XR Cell Viability Analyzer (Beckman Coulter, catalog number: 1096573) or Hemocytometer
- Operating Scissors (Fisher, catalog number: 13-806-4)
- Exel International Sterile disposable scalpels with surgical steel blades size 11 (Fisher, catalog number: 14-840-01)
- Dissecting forceps, fine tip, curved and straight (Fisher, catalog numbers: 08-875 and 08-880)
- Biological safety cabinet
- Surgical Autoclip System Kit (clip applier and remover are included) (Fine Science Tools, catalog number: 12120-00)
- Ear tags (1005-1) and Applicator (1005s1) (National Band & Tag Company)
- Digital Caliper 150 mm/6 (World Precision Instruments, catalog number: 501601 )
- E·Z Anesthesia (Euthanex CORP., Serial NO: EZV2007 )
Software
- Olympus cellSens Standard 1.17 image analysis software (Olympus, Center Valley, PA)
- SAS9.4 (SAS Institute Inc., Cary, NC, USA)
- R (Comprehensive R Archive Network, http://cran.us.r-project.org, 1997)
Procedure
- Prepare cells
- Seed cells into 10 cm dishes (0.5 x 106 cells/dish) three days before implantation.
- Change media (DMEM/F12 + 10% FBS + 1% Pen/Strep/Glutamine) the day before implantation.
- Harvest cells by trypsinization, rinse with PBS three times, count and resuspend cells in PBS at 2 x 107/ml.
Note: Total volume of cells prepared will depend on the number of mice needed. - Prepare 1.5 ml Eppendorf tube per mouse and mix 60 µl cell suspension in PBS with 60 µl Matrigel (the ratio of cells:Matrigel is 1:1).
Note: Since matrigel forms a gel above 10 °C, matrigel solution should be thawed overnight at 4 °C and kept on ice throughout the preparation. Tubes and tips for transferring Matrigel solution should also be chilled prior to implantation.
- Implant cells and Matrigel mixture into NOD.CB17-Prkdcscid/J (#00 1303 ; The Jackson Laboratory)
- Shave hair from the lateral body wall area.
- Administer analgesic (Buprenorphine HCl, diluted in sterile saline, 0.1 mg/kg SQ) 20-30 min before mouse surgery.
Note: Buprenorphin HCl is a controlled substance and should be stored in a double locked cabinet. - Anesthetize mouse by isoflurane inhalation until the mouse is unresponsive to toe pinch. Keep mouse warm and monitor breathing during procedure.
Note: You should follow the IACUC protocol to administer the right dose of isoflurane. - Disinfect surgical site with providone-iodine alternating with 70% ethanol for 3 times.
Note: See Video 1 for Steps B5-B9.
- Lift the skin with sterilized dissecting forceps and make ~5 mm incision with operating scissors in the lateral body wall just posterior to the axillary area.
- Make a subcutaneous pouch by blunt dissection.
- (Optional) Transfer 100 µl of cells mixed with Matrigel (1 x 106 cells) into the pouch using pipette with 200 µl wide bore Pipet tip to minimize the shearing of the cells.
- Place ear tag on mice.
- Close incision using clip applier with 9 mm surgical clips and place mouse in recovery cage. Mouse should recover from anesthesia within 1-2 min.Video 1. Subcutaneous implantation of cells and Matrigel mixture into mouse. (Animal study was performed according to the procedure approved by the University of Kentucky Institutional Animal Care and Use Committee (IACUC).)
- Monitor tumor growth
- Check clips and mouse health daily until clips are removed using clip remover from mouse; wound clips should be removed 7-10 days after surgery.
- Measure tumor size using a caliper 3 times/week and weigh mouse until tumor size reaches around 1,000-1,500 mm3 (Note: endpoint will depend on the goal of the experiments). Three parameters of the tumor: the length (the longest diameter), the width (the diameter perpendicular to the length), and the height were recorded.
- Harvest primary tumor tissues and organs
- Perform CO2 euthanasia and maintain CO2 flow for a minimum of 1 min after respiration ceases; then perform cervical dislocation.
Note: You should follow your IACUC protocol to sacrifice mouse. - Clean tumor area with 70% ethanol.
- Cut skin and remove tumor carefully and place tumor in a sterile tube containing DMEM/F12 media and keep it on ice.
- Perform necropsy to remove lungs or other organs such as liver, spleen, kidney or brain, and put them in tube containing DMEM/F12 media and keep the tube on ice.
- Rinse tissues with PBS.
- Fix tissues in 10% neutral buffered formalin overnight at 4 °C and replace solution with 70% ethanol before processing for paraffin embedding.
Note: Histopathology Facility may be required to process the samples for this step. - Stain tissues with hematoxylin and eosin (H&E).
- Examine H&E slides to identify foci of adenocarcinoma (for A549 cells) (Figure 1). These can be identified by recognizing characteristic architectural and cytologic features (e.g., glandular tumor formations, papillary growth, or lepidic tumor spread; loss of nuclear polarity, nuclei with fine chromatin and prominent nucleoli, anisonucleosis).
- Measure the diameter of each tumor foci in millimeters using a digital micrometer.
Note: We used the Olympus cellSens Standard 1.17 image analysis software.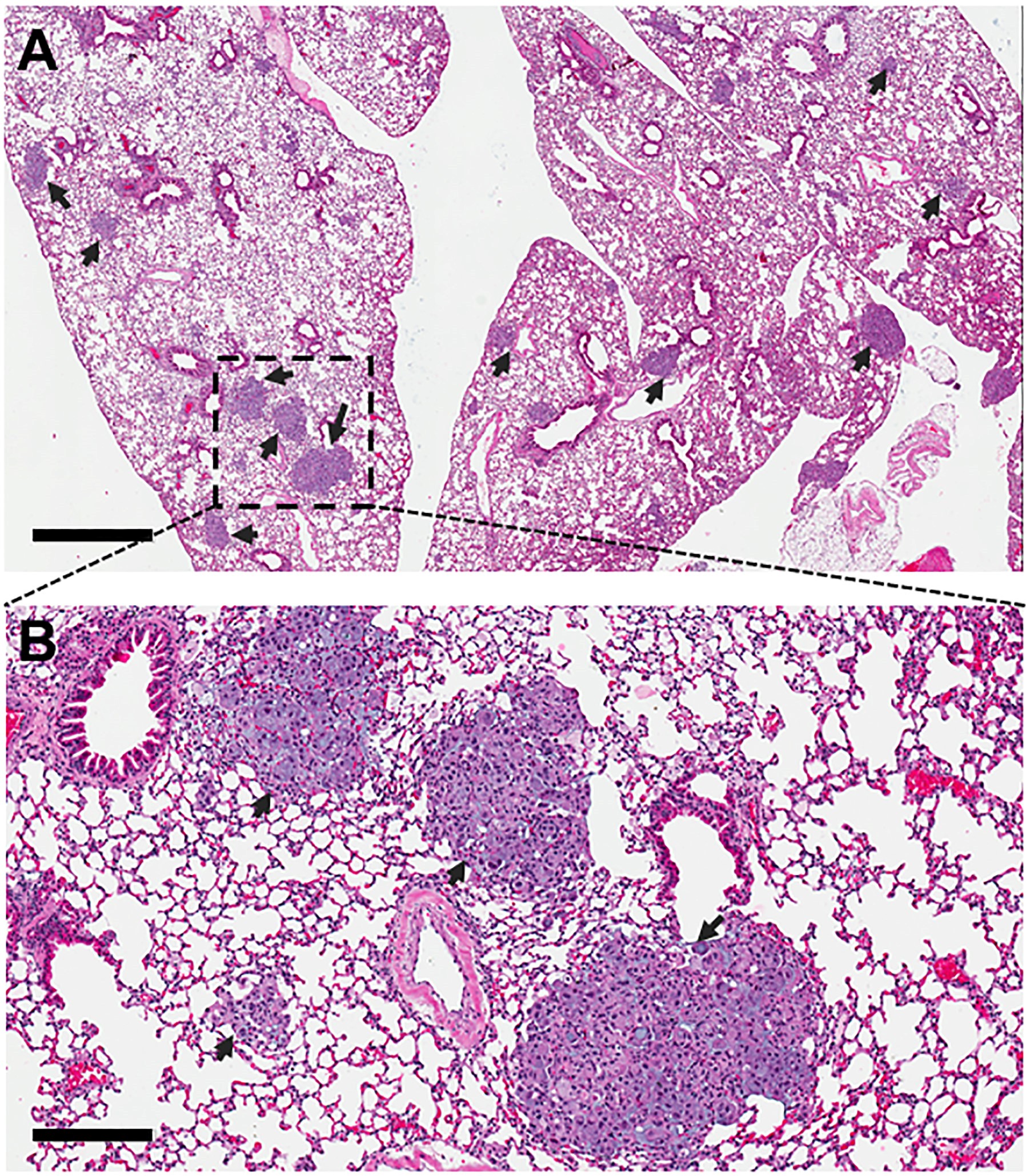
Figure 1. H&E staining of lung section. Magnification = 40x for image in (A) and 200x for image in (B). Scales bar = 1 mm in (A) and 100 µm in (B). Arrows indicate tumor foci. Representative images are shown.
- Perform CO2 euthanasia and maintain CO2 flow for a minimum of 1 min after respiration ceases; then perform cervical dislocation.
Data analysis
- Record total number of foci and calculate the tumor foci incidence (total number of foci/observation time (day)).
- Calculate tumor volume (V) based on the measurement from Step C2 as V = (L x W x H)/2 where V = tumor volume, L = tumor length, W = tumor width, and H = tumor height.
- Data analysis: For in vivo experimental metastasis study, a linear mixed effect model is used to fit the longitudinal tumor volumes and test the significant difference of mean tumor volume profiles between treatment groups (Figure 2). The Welch two-sample t-test should be used to test the difference of the mean tumor foci size between groups. A Poisson model is used to model the number of tumor foci incidence per day and significant difference of tumor foci incidence rates between groups is tested using Poisson test. Statistical difference between groups will be considered significant at the level of 0.05.
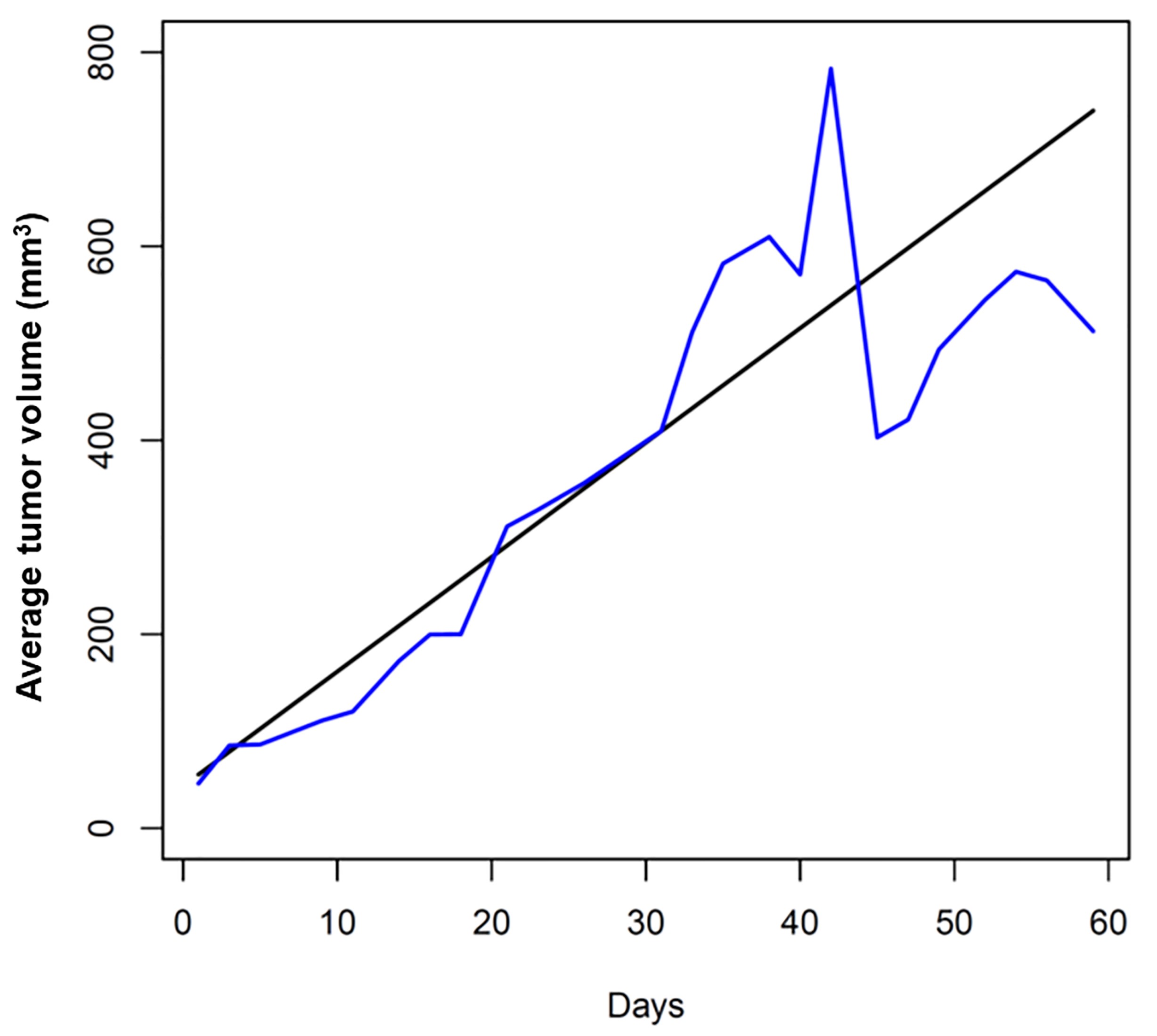
Figure 2. Tumor growth rate. Tumor growth curve was fitted and plotted using R software. The black line is fitted tumor volume profile and the blue line is the observed mean tumor volume.
Notes
- Tumor tissues and organs from necropsy can be rinsed with PBS and fixed immediately or temporary stored in media without serum for less than 30 min on ice before further processing.
- Assistance from a pathologist may be needed for slides assessment.
Acknowledgments
This study was supported by American Cancer Society Institutional Research Grant IRG-85-001-25 (MC); the Kentucky Lung Cancer Research program (MC); the National Center for Advancing Translational Sciences National Institutes of Health UL1TR000117 (MC); the University of Kentucky Center for Cancer and Metabolism [P20GM121327] for providing services through the Imaging Core and for the pilot funding [no grant number, to MC]); and the National Institutes of Health R01 CA109136 (KLO). The Markey Cancer Center Biospecimen Procurement and Translational Pathology, the Biostatistics and Bioinformatics and Cancer Research Informatics Shared Resource Facilities are supported by National Institutes of Health [P30 CA177558]. This protocol was briefly described and originally published in Liu et al., 2019.
Competing interests
The authors report no conflict of interest.
Ethics
Animal study was performed according to the procedure approved by the University of Kentucky Institutional Animal Care and Use Committee (IACUC).
This protocol was modified from Liu et al., 2019 and Morton et al., 2007.
References
- D'Antonio, C., Passaro, A., Gori, B., Del Signore, E., Migliorino, M. R., Ricciardi, S., Fulvi, A. and de Marinis, F. (2014). Bone and brain metastasis in lung cancer: recent advances in therapeutic strategies. Ther Adv Med Oncol 6(3): 101-114.
- Dutt, A. and Wong, K. K. (2006). Mouse models of lung cancer. Clin Cancer Res 12(14 Pt 2): 4396s-4402s.
- Gomez-Cuadrado, L., Tracey, N., Ma, R., Qian, B. and Brunton, V. G. (2017). Mouse models of metastasis: progress and prospects. Dis Model Mech 10(9): 1061-1074.
- Heist, R. S. and Engelman, J. A. (2012). SnapShot: non-small cell lung cancer. Cancer Cell 21(3): 448 e442.
- Justilien, V. and Fields, A. P. (2013). Utility and applications of orthotopic models of human non-small cell lung cancer (NSCLC) for the evaluation of novel and emerging cancer therapeutics. Curr Protoc Pharmacol 62: Unit 14 27.
- Khanna, C. and Hunter, K. (2005). Modeling metastasis in vivo. Carcinogenesis 26(3): 513-523.
- Liu, L., Qi, L., Knifley, T., Piecoro, D. W., Rychahou, P., Liu, J., Mitov, M. I., Martin, J., Wang, C., Wu, J., Weiss, H. L., Butterfield, D. A., Evers, B. M., O'Connor, K. L. and Chen, M. (2019). S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J Biol Chem 294(18): 7516-7527.
- Morton, C. L. and Houghton, P. J. (2007). Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc 2(2): 247-250.
- Okada, S., Vaeteewoottacharn, K. and Kariya, R. (2019). Application of highly immunocompromised mice for the establishment of patient-derived xenograft (PDX) models. Cells 8(8).
- Steeg, P. S. (2016). Targeting metastasis. Nat Rev Cancer 16(4): 201-218.
- Whitsett, T. G., Inge, L. J., Dhruv, H. D., Cheung, P. Y., Weiss, G. J., Bremner, R. M., Winkles, J. A. and Tran, N. L. (2013). Molecular determinants of lung cancer metastasis to the central nervous system. Transl Lung Cancer Res 2(4): 273-283.
- Zheng, S., El-Naggar, A. K., Kim, E. S., Kurie, J. M. and Lozano, G. (2007). A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene 26(48): 6896-6904.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Qi, L., Knifley, T., Piecoro, D. W., Rychahou, P., Wu, J., O'Connor, K. L. and Chen, M. (2020). In vivo Tumor Growth and Spontaneous Metastasis Assays Using A549 Lung Cancer Cells. Bio-protocol 10(7): e3579. DOI: 10.21769/BioProtoc.3579.
- Liu, L., Qi, L., Knifley, T., Piecoro, D. W., Rychahou, P., Liu, J., Mitov, M. I., Martin, J., Wang, C., Wu, J., Weiss, H. L., Butterfield, D. A., Evers, B. M., O'Connor, K. L. and Chen, M. (2019). S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J Biol Chem 294(18): 7516-7527.
Category
Cancer Biology > Invasion & metastasis > Animal models > Cell isolation and culture
Cell Biology > Cell viability > Cell proliferation
Cancer Biology > General technique > Animal models > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link