- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Flow Cytometry Measurement of Glucocerebrosidase Activity in Human Monocytes
Published: Vol 10, Iss 7, Apr 5, 2020 DOI: 10.21769/BioProtoc.3572 Views: 6066
Reviewed by: Maria NguyenVemika ChandraSébastien Gillotin

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1413 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1441 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 287 Views
Abstract
Glucocerebrosidase (GCase) is an important enzyme for the metabolism of glycolipids. GCase enzyme deficiency is implicated in human disease and the efficient measurement of GCase activity is important for evaluating the efficacy of therapeutics targeting this enzyme. Existing approaches to measure GCase activity include whole blood mass spectrometry-based assays, where an internal standard is used to measure the accumulation of ceramide following metabolism of the synthetic substrate C12-glucocerebroside, and the utilisation of fluorescent probes that bind active GCase and/or release fluorescent metabolites upon cleavage by GCase. Here, we describe the application of a fluorescence-activated cell sorter-based assay to efficiently quantitate GCase enzyme activity in the monocyte population of human peripheral blood mononuclear cells. The cell-permeable GCase substrate 5-(Pentafluorobenzoylamino) Fluorescein Di-beta-D-Glucopyranoside (PFB-FDGlu) provides a means to measure GCase activity, whereby enzymatic cleavage yields the green-fluorescent PFB-F dye, detectable in the FL-1 channel of a flow cytometer. An inhibitor of lysosomal GCase activity, conduritol B-epoxide, is employed to ensure specificity. This protocol provides an advantageous approach for measuring GCase activity in living individual cells.
Keywords: GlucocerebrosidaseBackground
Glucocerebrosidase (GCase), encoded by the GBA1 gene, is a lysosomal hydrolase that converts glucosylceramide to glucose and ceramide. Loss of function homozygous, and compound heterozygous missense mutations in GBA1 are associated with the accumulation of glucosylceramide and the lysosomal storage disorder Gaucher’s disease (Sidransky, 2012). Whereas heterozygous mutations in GBA1 are associated with increased risk of developing the neurodegenerative movement disorder Parkinson’s disease (PD) (Sidransky and Lopez, 2012). While GBA1 mutations occur in approximately 5-15% of PD cases (Neumann et al., 2009; Sidransky et al., 2009; Lesage et al., 2011), reduced GCase activity is also observed in PD patients without GBA1 mutations (Murphy et al., 2014; Alcalay et al., 2015; Atashrazm et al., 2018), and there is increasing evidence supporting the notion that restoring GCase function is a valid therapeutic option for PD (Schapira 2015; Sardi et al., 2018). Subsequently, a number of small molecule chaperones have been described that can restore GCase function (Khanna et al., 2010 and 2012; Patnaik et al., 2012; McNeill et al., 2014).
The assessment of GCase activity in PD patients can help inform which patients may benefit from such therapies, by identifying those with reduced enzyme activity. Assessment of GCase activity could also be used to gauge the efficacy of proposed therapies to restore GCase function. The efficacy of chaperone-based therapies for restoring GCase function have predominantly been assessed by measuring changes in GCase protein by immunoblotting, or by indirectly measuring GCase activity using mass spectrometry-based assays. Although these approaches have proven useful and do have advantages (Wolf et al., 2018), they are also limited in their ability to distinguish between blood cell types, assay GCase activity in intact living cells, and are potentially less clinically amenable than other techniques. Another current approach to measuring GCase activity involves the use of the non-natural substrate 4-MUG (4-Methylumbelliferyl β-D-galactopyranoside), which when cleaved by GCase produces the fluorescent product 4-methylumbelliferon (4-MU). However, 4-MUG is also a substrate for non-lysosomal GCase (encoded by GBA2) (Boot et al., 2007) requiring the purification of lysosomes or performing the assay at distinguishing pH to add specificity.
The current protocol presented here utilises a selective lysosomal GCase substrate 5-(Pentafluorobenzoylamino) Fluorescein Di-beta-D-Glucopyranoside (PFB-FDGlu), which is metabolised by GCase to yield fluorescein. PFB-FDGlu is cell permeable and can be used with a flow cytometer to measure GCase activity in living cells on a single-cell basis. This protocol has been adapted from earlier studies describing and validating the use of PFB-FDGlu for assessing GCase activity, primarily in the context of Gaucher’s disease (Lorincz et al., 1997; van Es et al., 1997; Rudensky et al., 2003). The ability to combine PFB-FDGlu with other phenotypic indicators, such as a viability stain and antibodies to label cell surface markers is advantageous, as it enables identification of distinct target cell populations in which GCase activity may be modulated.
Materials and Reagents
- Ethanol wipes (Livingstone, catalog number: LWMS )
- Plasters (Livingstone, catalog number: ASP23N )
- Needles 23G (BD Biosciences, catalog number: 367288 )
- BD vacutainer one-use holders (BD Biosciences, catalog number: 364815 )
- BD CPT sodium citrate Vacutainer Glass Cell Preparation Tubes (BD Biosciences, catalog number: 362782 )
- 10 μl XL Biotix Filter Tip (Interpath, catalog number: M00119FC )
- 100 μl Aerosol Barrier Tip (Interpath, catalog number: 24600 )
- 1,000 μl Aerosol Barrier Tip (Interpath, catalog number: 24800 )
- Falcon 5 ml round bottom polystyrene test tube with cell strainer cap, 12 x 75 mm (In Vitro Technologies, catalog number: FAL352235 )
- 1.5 ml EppendorfTM Safe-Lock microcentrifuge tubes (Sigma-Aldrich, catalog number: T9661 )
- Countess cell counting chamber slides (Life Technologies, catalog number: C10312 )
- Sterile syringe filter 0.22 µm (Millipore, catalog number: SLGVV255F )
- 1.5 ml and 15 ml centrifuge tubes
- PPE (lab coat, safety glasses, disposable gloves)
- MilliQ water
- Ice box and ice
- RPMI 1640 Medium (Thermo Fisher Scientific, catalog number: 1187093 )
- Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, catalog number: 10100147 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2438 )
- Penicillin-Streptomycin (100x) (Thermo Fisher Scientific, catalog number: 15140122 )
- L-Glutamine (Thermo Fisher Scientific, catalog number: 25030081 )
- CD3 for T-lymphocytes (BD Biosciences, catalog number: 560365 )
- CD19 for B-lymphocytes (BD Biosciences, catalog number: 555415 )
- CD16 for non-classical monocytes (BD Biosciences, catalog number: 560475 )
- 5-(Pentafluorobenzoylamino) Fluorescein Di-beta-D-Glucopyranoside (PFB-FDGlu) (Thermo Fisher Scientific, catalog number: P11947 )
- PE-Cy7 Mouse Anti-Human CD14 antibody (BD Biosciences, catalog number: 557742 )
- PE-Cy7 Mouse IgG2a, k Isotype Control (BD Biosciences, catalog number: 557907 )
- 1x DPBS (Ca2+/Mg2+ free) (Thermo Fisher Scientific, catalog number: 14190144 )
- 1 M HEPES (Sigma-Aldrich, catalog number: H0887 )
- Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) (Merck, catalog number: E5134 )
- Sodium Hydroxide (NaOH) (Merck, catalog number: 221465 )
- Trypan blue 0.4% (Thermo Fisher Scientific, catalog number: T10282 )
- Culture media (500 ml) (see Recipes)
- 0.1 M EDTA solution (500 ml) (see Recipes)
- Fluorescence-activated cell sorting (FACS) Buffer (500 ml) (see Recipes)
- CBE 50 mM stock solution (see Recipes) (Sigma-Aldrich, catalog number: C5424-5MG )
- PFB-FDGlu 37.5 mM stock solution (see Recipes)
Equipment
- Tube rack to hold 1.5, 5, 15 ml tubes
- Class II Biosafety cabinet
- Micropipettes (P10, P100, P1000)
- S1 Pipette Filler (Thermo Fisher Scientific, catalog number: 9521 )
- Heraeus Multifuge X1 Centrifuge with inserts for 15 ml conical tubes (Thermo Fisher Scientific)
- Centrifuge (Eppendorf, model: 5415 R )
- Countess II FL automated cell counter (Thermo Fisher Scientific)
- Hotplate magnetic stirrer
- Magnetic stir bar
- Magnetic stirring pad
- pH meter
- 4 °C refrigerator
- -20 °C freezer
- 37 °C 5% CO2 incubator
- Vortex
- Flow cytometer equipped with a 488 nm laser and 530 nm-FITC filter (i.e., BD LSR Fortessa X-20)
- Computer for analysis
Software
- BD FACSDivaTM software (BD Biosciences)
- FlowJo software (Tree Star, OR, USA)
- GraphPad Prism software (GraphPad Software Inc., CA, USA)
Procedure
- Aseptic technique
Conduct experiments in a class II biological safety cabinet using appropriate PPE to maintain safety and sterility. - Isolation of human peripheral blood mononuclear cells (PBMC)
- Perform venepuncture with the arm in a downward position. Collect venous blood into 8 ml BD Vacutainer Cell Preparation Tubes (CPT), keeping tubes upright following blood collection. Immediately prior to centrifugation, invert tubes 8 times to remix the blood sample.
- Centrifuge at 1,800 x g for 20 min at room temperature (21 °C) in a swing bucket centrifuge with acceleration and deceleration set at 9.
- Collect PBMC layer with sterile glass pipette (whitish layer just beneath the plasma) and transfer to a 15 ml tube. For more specific details and images on collecting the PBMC layer from CPT tubes please refer to the following Bio-protocol publication https://bio-protocol.org/e2103 (Puleo et al., 2017).
- Add room temperature 1x DPBS to bring volume to 15 ml and invert tube 5 times, followed by centrifugation for 15 min at 300 x g at room temperature.
- Aspirate supernatant and resuspend pellet in 10 ml of warmed (37 °C) culture media for cell counting.
- Dilute PBMC suspension 1:1 in Trypan blue, transfer mixture to a cell counting chamber slide, and count PBMCs using an automated cell counter.
- Determine the required volume to obtain 1 x 106 cells.
- Distribution of PBMCs into Eppendorf tubes
- Add 1 x 106 PBMCs per Eppendorf tube, with a total of three Eppendorf tubes for each subject, as shown in Table 1. One tube receives CBE (tube 1) and one tube receives DMSO (tube 2), with each of these followed by the addition of PFB-FDGlu substrate. The third tube is an isotype control.
Table 1. Distribution of PBMCs into Eppendorf tubes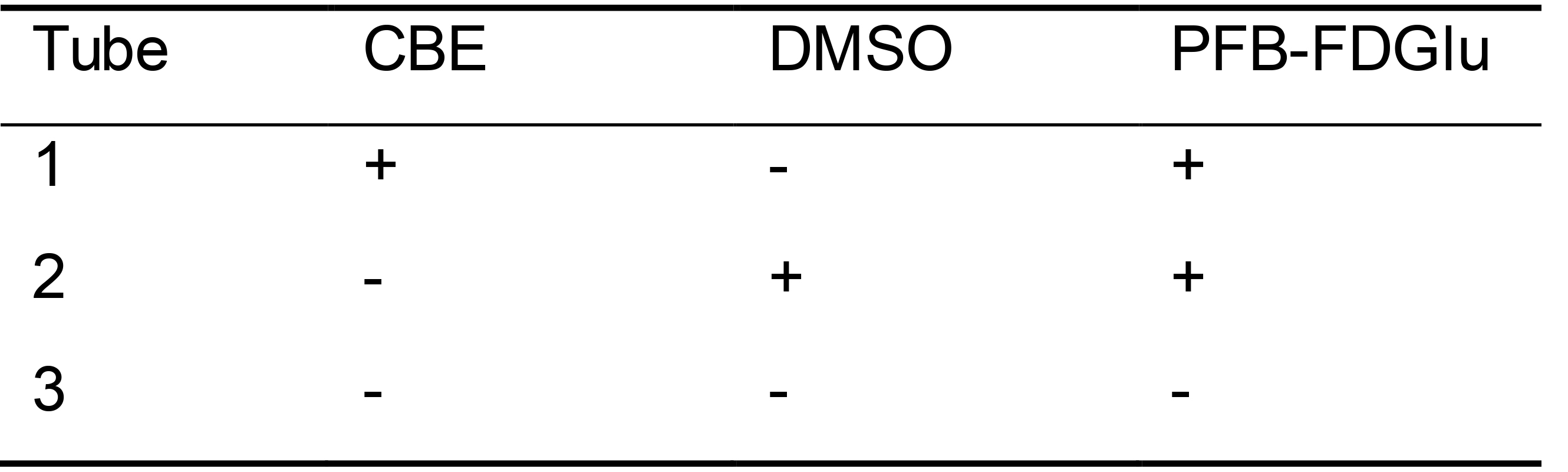
- Add 1 x 106 PBMCs per Eppendorf tube, with a total of three Eppendorf tubes for each subject, as shown in Table 1. One tube receives CBE (tube 1) and one tube receives DMSO (tube 2), with each of these followed by the addition of PFB-FDGlu substrate. The third tube is an isotype control.
- Addition of CBE or DMSO to appropriate tubes
- Centrifuge tubes at 300 x g for 5 min at room temperature and decant supernatant.
- Gently resuspend cells in 96 μl of culture media.
- Add 2 μl of 50 mM CBE stock solution into tube 1 to achieve a final concentration of 1 mM, and add 2 μl of DMSO into tube 2
- Vortex tubes gently to mix.
- Incubate at 37 °C with 5% CO2 for 60 min.
- Addition of PFB-FDGlu substrate to appropriate tubes
- Following the 60 min incubation, add 2 μl of 37.5 mM PFB-FDGlu stock solution into tubes 1 and 2 to achieve a final concentration of 0.75 mM.
- Vortex tubes gently to mix.
- Incubate at 37 °C with 5% CO2 for 30 min.
- Termination of reaction
- To terminate the reaction, add 1 ml of ice-cold FACS buffer.
- Pellet cells via centrifugation at 300 x g for 5 min at 4 °C and decant supernatant.
- Staining for CD14 positive monocytes
- Resuspend cells in 95 μl of ice-cold flow buffer and add 5 μl of anti-CD14 antibody into tubes 1 and 2.
- Add 5 μl of isotype control antibody into tube 3 if using BD. For other brands of isotype control, ensure usage at the same concentration as the BD anti-CD14 antibody.
- Vortex tubes gently to mix.
- Incubate for 20 min at 4 °C, protected from light.
- Add 1 ml of FACS buffer to tubes, centrifuge at 300 x g for 5 min at 4 °C, and discard supernatant.
- Resuspend cells in 350 μl FACS buffer.
- Pass each cell suspension through the cell strainer cap into the 5 ml polypropylene tube compatible with the flow cytometer.
- Store tubes shielded from light at 4 °C until flow cytometry data acquisition.
- Data acquisition
- Acquire data on a BD LSR Fortessa X-20 Cell Analyser using FACSDiva or equivalent cytometer and software, collecting a minimum of 50,000 events per tube.
- Export FCS files from FACSDiva software.
Data analysis
- Gating strategy
- Analyse FCS files using FlowJo software. To remain objective, create the gates described below using tube 3 (isotype control tube), and then apply the gates to tubes 1 and 2 by selecting and dragging the gate series to the “All Samples” tab under the “Group” tab.
- Firstly, perform doublet and debris exclusion by plotting area against height for forward scatter (FSC: Figure 1A), and then for side scatter (SSC: Figure 1B), to ensure only single cells are included in the analysis. Label these “singlets FSC” and “singlets SSC”, respectively.
- Use FSC (size) and SSC (granularity) parameters to gate the mononuclear cell population (Figure 1C), and label this “mononuclear cells”.
- Next, plot CD14-PE-Cy7 fluorescence against SSC-A and select the CD14-positive population (Figure 1D), from which GCase activity will be measured. The isotype negative control tube can be used to identify and eliminate non-specific binding of the antibody, as any signal above isotype control signal can be interpreted as true CD14 positive fluorescence. Label the CD14 positive population as “CD14+ monocytes”.

Figure 1. Representative biaxial flow cytometry plots depicting the gating strategy used to assess GCase activity in live monocytes. Events were acquired using a BD LSR Fortessa Cell Analyser. First, two singlet gates were made, using FSC (A) and SSC (B), followed by gating on PBMCs (C), and then CD14+ monocytes (D). The median fluorescence intensity resulting from PFB-FDGlu metabolism within the monocyte population was then quantified (E).
- Calculating GCase activity
Express GCase activity as an index, obtained by dividing the median fluorescence intensity of cells treated without CBE by the median fluorescence intensity of cells treated with CBE. To find the median fluorescence intensity resulting from PFB-FDGlu metabolism in each tube, right-click on the “CD14+ monocytes” tab under the tube name, click “Add Statistic”, and then co-select “Median” and “FL1-A” (or equivalent channel used to detect PFB-FDGlu fluorescence). Click “Add”, and a tab displaying the median fluorescence intensity of PFB-FDGlu metabolism will appear under the “CD14+ monocytes” tab. - Statistical analysis
Perform statistical analyses using GraphPad Prism software to compare the GCase activity index between groups of interest. As an example, in our original research article (Atashrazm et al., 2018) analyses were performed to compare GCase activity between PD patients (n = 48) and healthy controls (n = 44) and this data can be found in Figure 3F.
Notes
- This assay may also be performed on PBMCs which have been cryopreserved. We recommend a recovery incubation period of ~2 h upon thawing, prior to beginning the assay.
- We have experienced large variation with different lots of PFB-FDGlu and recommend using a single lot number per experiment.
- We have also experienced a decline in PFB-FDGlu performance with prolonged storage after reconstitution. We recommend using reconstituted PFB-FDGlu within 1 week.
Recipes
- Culture media (500 ml)
- Combine 440 ml of RPMI 1640 with 5 ml of penicillin/streptomycin 100x, 5 ml of L-Glutamine, and 50 ml of fetal bovine serum
- Filter sterilize and store at 4 °C for up to one month
- 0.1 M EDTA solution (500 ml)
- Combine 18.61 g of EDTA with 500 ml MilliQ Water, using a magnetic stirring pad with heating to dissolve
- Adjust pH to reach 8.0 using sodium hydroxide (NaOH)
- Autoclave at 125 °C for 25 min to sterilize
- Store at room temperate for several months
- Fluorescence-activated cell sorting (FACS) Buffer (500 ml)
- Combine DPBS (Ca2+/Mg2+ free) with 1 mM EDTA, 25 mM HEPES, and 5% v/v FBS to produce FACS buffer
- Filter sterilize and store at 4 °C for several months
- CBE 50 mM stock solution
- Bring one 5 mg vial of CBE to room temperature and aseptically combine with 616.9 μl DMSO to achieve a stock solution of 50 mM
- Store as single use aliquots at -20 °C for several months
- PFB-FDGlu 37.5 mM stock solution
- Bring one 5 mg vial of PFB-FDGlu to room temperature and aseptically combine with 154 μl dimethyl sulfoxide (DMSO) to achieve a stock solution of 37.5 mM
- Store as single use aliquots at -20 °C for no longer than one week
Acknowledgments
This protocol is derived from a published study (Atashrazm et al., 2018) which was jointly funded by the Michael J Fox Foundation for Parkinson’s disease research and the Shake it up Australia Foundation. It is important to note that this derived protocol was itself adapted from earlier pioneering studies establishing the use of PFB-FDGlu to asses GCase activity (Lorincz et al., 1997; van Es et al., 1997; Rudensky et al., 2003).
Competing interests
The authors have no financial or non-financial competing interests to declare.
Ethics
All human blood samples were obtained with informed consent. Experiments were conducted in accordance with relevant guidelines and regulations and were approved by the University of Sydney Human Research Ethics Committee (#2016/363).
References
- Alcalay, R. N., Levy, O. A., Waters, C. C., Fahn, S., Ford, B., Kuo, S. H., Mazzoni, P., Pauciulo, M. W., Nichols, W. C., Gan-Or, Z., Rouleau, G. A., Chung, W. K., Wolf, P., Oliva, P., Keutzer, J., Marder, K. and Zhang, X. (2015). Glucocerebrosidase activity in Parkinson's disease with and without GBA mutations. Brain 138(Pt 9): 2648-2658.
- Atashrazm, F., Hammond, D., Perera, G., Dobson-Stone, C., Mueller, N., Pickford, R., Kim, W. S., Kwok, J. B., Lewis, S. J. G., Halliday, G. M. and Dzamko, N. (2018). Reduced glucocerebrosidase activity in monocytes from patients with Parkinson's disease. Sci Rep 8(1): 15446.
- Boot, R. G., Verhoek, M., Donker-Koopman, W., Strijland, A., van Marle, J., Overkleeft, H. S., Wennekes, T. and Aerts, J. M. (2007). Identification of the non-lysosomal glucosylceramidase as beta-glucosidase 2. J Biol Chem 282(2): 1305-1312.
- Khanna, R., Benjamin, E. R., Pellegrino, L., Schilling, A., Rigat, B. A., Soska, R., Nafar, H., Ranes, B. E., Feng, J., Lun, Y., Powe, A. C., Palling, D. J., Wustman, B. A., Schiffmann, R., Mahuran, D. J., Lockhart, D. J. and Valenzano, K. J. (2010). The pharmacological chaperone isofagomine increases the activity of the Gaucher disease L444P mutant form of beta-glucosidase. FEBS J 277(7): 1618-1638.
- Khanna, R., Pellegrino, L., Soska, R., Lun, Y., Feng, J., Frascella, M., E.Ranes, B., Guillen, D., Garcia, A., J.Flanagan, J., J.Lockhart, D., W.Clark, S. and J.Valenzano, K. (2012). Exploring the use of pharmacological chaperone at3375 alone and in combination with recombinant human ß-glucosidase for gaucher dIsease. Mol Genet Metab 105(2): S40.
- Lesage, S., Anheim, M., Condroyer, C., Pollak, P., Durif, F., Dupuits, C., Viallet, F., Lohmann, E., Corvol, J. C., Honore, A., Rivaud, S., Vidailhet, M., Durr, A., Brice, A. and French Parkinson's Disease Genetics Study, G. (2011). Large-scale screening of the Gaucher's disease-related glucocerebrosidase gene in Europeans with Parkinson's disease. Hum Mol Genet 20(1): 202-210.
- Lorincz, M., Herzenberg, L. A., Diwu, Z., Barranger, J. A. and Kerr, W. G. (1997). Detection and isolation of gene-corrected cells in Gaucher disease via a fluorescence-activated cell sorter assay for lysosomal glucocerebrosidase activity. Blood 89(9): 3412-3420.
- McNeill, A., Magalhaes, J., Shen, C., Chau, K. Y., Hughes, D., Mehta, A., Foltynie, T., Cooper, J. M., Abramov, A. Y., Gegg, M. and Schapira, A. H. (2014). Ambroxol improves lysosomal biochemistry in glucocerebrosidase mutation-linked Parkinson disease cells. Brain 137(Pt 5): 1481-1495.
- Murphy, K. E., Gysbers, A. M., Abbott, S. K., Tayebi, N., Kim, W. S., Sidransky, E., Cooper, A., Garner, B. and Halliday, G. M. (2014). Reduced glucocerebrosidase is associated with increased alpha-synuclein in sporadic Parkinson's disease. Brain 137(Pt 3): 834-848.
- Neumann, J., Bras, J., Deas, E., O'Sullivan, S. S., Parkkinen, L., Lachmann, R. H., Li, A., Holton, J., Guerreiro, R., Paudel, R., Segarane, B., Singleton, A., Lees, A., Hardy, J., Houlden, H., Revesz, T. and Wood, N. W. (2009). Glucocerebrosidase mutations in clinical and pathologically proven Parkinson's disease. Brain 132(Pt 7): 1783-1794.
- Patnaik, S., Zheng, W., Choi, J. H., Motabar, O., Southall, N., Westbroek, W., Lea, W. A., Velayati, A., Goldin, E., Sidransky, E., Leister, W. and Marugan, J. J. (2012). Discovery, structure-activity relationship, and biological evaluation of noninhibitory small molecule chaperones of glucocerebrosidase. J Med Chem 55(12): 5734-5748.
- Puleo, A., Carroll, C., Maecker, H.T. and Gupta, R. (2017). Isolation of PBMCs using Vacutainer Cellular Preparation Tubes (CPT). Bio-protocol 7(2): e2103.
- Rudensky, B., Paz, E., Altarescu, G., Raveh, D., Elstein, D. and Zimran, A. (2003). Fluorescent flow cytometric assay: a new diagnostic tool for measuring beta-glucocerebrosidase activity in Gaucher disease. Blood Cells Mol Dis 30(1): 97-99.
- Sardi, S. P., Cedarbaum, J. M. and Brundin, P. (2018). Targeted therapies for parkinson's disease: from genetics to the clinic. Mov Disord 33(5): 684-696.
- Schapira, A. H. (2015). Glucocerebrosidase and Parkinson disease: Recent advances. Mol Cell Neurosci 66(Pt A): 37-42.
- Sidransky, E. (2012). Gaucher disease: insights from a rare Mendelian disorder. Discov Med 14(77): 273-281.
- Sidransky, E. and Lopez, G. (2012). The link between the GBA gene and parkinsonism. Lancet Neurol 11(11): 986-998.
- Sidransky, E., Nalls, M. A., Aasly, J. O., Aharon-Peretz, J., Annesi, G., Barbosa, E. R., Bar-Shira, A., Berg, D., Bras, J., Brice, A., Chen, C. M., Clark, L. N., Condroyer, C., De Marco, E. V., Durr, A., Eblan, M. J., Fahn, S., Farrer, M. J., Fung, H. C., Gan-Or, Z., Gasser, T., Gershoni-Baruch, R., Giladi, N., Griffith, A., Gurevich, T., Januario, C., Kropp, P., Lang, A. E., Lee-Chen, G. J., Lesage, S., Marder, K., Mata, I. F., Mirelman, A., Mitsui, J., Mizuta, I., Nicoletti, G., Oliveira, C., Ottman, R., Orr-Urtreger, A., Pereira, L. V., Quattrone, A., Rogaeva, E., Rolfs, A., Rosenbaum, H., Rozenberg, R., Samii, A., Samaddar, T., Schulte, C., Sharma, M., Singleton, A., Spitz, M., Tan, E. K., Tayebi, N., Toda, T., Troiano, A. R., Tsuji, S., Wittstock, M., Wolfsberg, T. G., Wu, Y. R., Zabetian, C. P., Zhao, Y. and Ziegler, S. G. (2009). Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med 361(17): 1651-1661.
- van Es, H. H., Veldwijk, M., Havenga, M. and Valerio, D. (1997). A flow cytometric assay for lysosomal glucocerebrosidase. Anal Biochem 247(2): 268-271.
- Wolf, P., Alcalay, R. N., Liong, C., Cullen, E., Pauciulo, M. W., Nichols, W. C., Gan-Or, Z., Chung, W. K., Faulkner, T., Bentis, C., Pomponio, R. J., Ma, X., Kate Zhang, X., Keutzer, J. M. and Oliva, P. (2018). Tandem mass spectrometry assay of beta-glucocerebrosidase activity in dried blood spots eliminates false positives detected in fluorescence assay. Mol Genet Metab 123(2): 135-139.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hughes, L. P., Halliday, G. M. and Dzamko, N. (2020). Flow Cytometry Measurement of Glucocerebrosidase Activity in Human Monocytes. Bio-protocol 10(7): e3572. DOI: 10.21769/BioProtoc.3572.
Category
Cell Biology > Cell-based analysis > Enzymatic assay
Biochemistry > Protein > Fluorescence
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









