- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single-cell qPCR Assay with Massively Parallel Microfluidic System
Published: Vol 10, Iss 6, Mar 20, 2020 DOI: 10.21769/BioProtoc.3563 Views: 5762
Reviewed by: Xi FengChaitali BasoleAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

DNA in situ Hybridizations for VEGFA Gene Locus (6p12) in Human Tumor Tissue
Mariacarla Andreozzi [...] Luigi Terracciano
Aug 5, 2015 9962 Views

Enhanced-ice-COLD-PCR for the Sensitive Detection of Rare DNA Methylation Patterns in Liquid Biopsies
Florence Mauger and Jörg Tost
Dec 5, 2019 6408 Views

In-Cell Western Protocol for Semi-High-Throughput Screening of Single Clones
Arpita S. Pal [...] Andrea L. Kasinski
Aug 20, 2022 3476 Views
Abstract
The single-cell transcriptome is the set of messenger RNA molecules expressed in one cell. It is extremely variable and changes according to external, physical and biochemical conditions. Due to sensitivity shortages, most of genetic studies use bulk samples, providing only the average gene expression. Single-cell technologies have provided a powerful approach to a more detailed understanding of the heterogenic populations and minority cells. However, since it is still a quite novel technique, standardized protocol has to be established. Single-cell qPCR, although partly limited by the number of genes, is relatively simple to analyze. Therefore, its use is accessible without the necessity to recourse to complex bioinformatics analyses. The main steps for single-cell qPCR, as illustrated in this protocol, are composed by single-cell isolation, cell lysate, cDNA reverse-transcription synthesis, amplification for cDNA library generation, and finally, quantitative polymerase chain reaction.
Keywords: Single-cellBackground
The single-cell transcriptome is the complete set of messenger RNA (mRNA) molecules expressed in one cell. It is extremely variable and changes according to external, physical and biochemical conditions. Hence, it is the source of the biological heterogeneity, in which the cells from the same environment are similar -but not identical- to the other ones.
Due to sensitivity shortages, most of genetic studies use bulk samples, with hundreds to thousands of cells, providing only the average gene expression. This vastly limits the study of minor populations of cells, which may poses specific properties, such as drug resistance or metastatic ability in the case of cancer. Single-cell technologies enable the analysis of the transcriptome at single cell level, unraveling the complexity of heterogenic populations. These technologies have been applied not only in cancer but in many cell biology studies, including adult tissues, stem cells, immune cells; as well as other fields such as microbiology and virology (Wen and Tang, 2016; Karaiskos et al., 2017; Rato et al., 2017; Woyke et al., 2017; Zheng et al., 2017).
There are two main single-cell transcriptome approaches: mRNA sequencing and quantitative polymerase chain reaction. Although mRNA sequencing enables the study of the whole transcriptome without prior knowledge, manipulating an extremely large amount of genes can be overwhelming for novel researchers in the field; moreover high levels of bioinformatics are required (Prieto-vila et al., 2018). To counterbalance, single-cell qPCR analysis is limited to a certain number of genes, but can be analyzed as normal qPCR, being more approachable.
In our previous research, we aimed to study a drug resistant subpopulation of cells by using single-cell qPCR in breast cancer cell lines. With that purpose, we used massively parallel single-cell amplification, that allowed us the study of 96 genes at single cell level to 96 cells per run (Prieto-Vila et al., 2019). This is one of the most commonly used techniques for single-cell qPCR in which reactions are carried in specific 96-well plates, where individual samples are reverse transcribed at the same time. The C1 nanofluidics along with the Fluidigm systems, are the most commonly used systems for sc-qPCR. C1 machine is a hydrodynamic cell trap chip, formed by a net of channels where the cells are eluted and single cells are fiscally separated their size. Thanks to this system, rarely doublets are found. The 96 single cells are then eluded into 96 individual wells. On these wells lysis, reverse transcription, and pre-amplification for qPCR are carried in parallel using a very small reaction volume (Ziegenhain et al., 2017). Following, massive parallel qPCR is done by analyzing the expression of 96 genes in the 96 individual cells in a matrix-manner chip by the Fluidigm system.
In the present protocol, we provide a single-cell transcriptome analysis protocol, including experimental details to perform single-cell qPCR with the Fluidigm systems.
Materials and Reagents
- Pipette tips 1,000 μl/200 μl/20 μl (Sorenson Bioscience, catalog numbers: 34000 / 14220 / 15020 )
- Low binding centrifuge tubes 1.5 ml (Watson BioLab, catalog number: pk-15 c-500 )
- Rainin LTS tips 10 μl (Rainin, catalog number: 30389228 )
- qPCR 96-well plates (Applied Biosystems, catalog number: 4346907 )
- Film for qPCR plates (Applied Biosystems, catalog number: 4311971 )
- Nuclease-Free water (Ambion, catalog number: AM9932 )
For cell culture
- Cell culture 100 mm dishes (Thermo Fisher Scientific, catalog number: 172931 )
- 15 ml conical centrifuge tubes (Thermo Fisher Scientific, catalog number: 339650 )
- Desired cell line, in this case we used the breast cancer cell lines MDA-MB-231 and MCF7
- RPMI 1640x basic medium (Thermo Fisher Scientific, catalog number: C11875500BT )
- Fetal bovine serum (FBS) (Gibco, catalog number: 10270-106 )
Note: FBS should be heat inactivated at 56 °C for 30 min before use. - Antibiotic/antimitotic solution (Thermo Fisher Scientific, catalog number: 15240062 )
- Phosphate buffer Saline (PBS) (DS Pharma Biomedical, catalog number: DSBN200 )
- TrypLE Express (Thermo Fisher Scientific, catalog number: 12604013 )
- Trypan Blue Solution 0.4% (Gibco, catalog number: 15250-061 )
- Neubauer Improved C-Chip (NanoEnTek, catalog number: DHC-N01 )
- 0.5 mol/L EDTA (Nacalai Tesque, catalog number: 06894-14 )
- Initial cell suspension buffer (see Recipes)
For single cell isolation and pre-amplification
- C1 Single-Cell Auto Prep IFC for Preamp (10-17 μm) (Fluidigm, catalog number: 100-5749 )
Note: There are several channel size plates to adjust to the cell size. See notes for more information. - 20x gene specific Assays (Taqman)
- C1 Single-Cell Auto Prep Reagent Kit (Fluidigm, catalog number: 100-5319 )
- Ambion single cell-to-Ct qRT-PCR Kit (Thermo Fisher Scientific, catalog number: 4458237 )
- Live/Dead Kit for mammalian cells (Life Technologies, catalog number: L-3224 ) (see Recipes)
- Lysis mix (see Recipes)
- Reverse transcription final mix (see Recipes)
- 0.2x Pooled Taqman primers (see Recipes)
- Pre-amplification mix (see Recipes)
For single cell qPCR
- Fluidigm 96.96 quantitative PCR Dynamic Array microfluidic chips (Fluidigm, catalog number: BMK-M-96.96GT )
- Taqman Universal PCR Master Mix 2x (Applied Biosystems, catalog number: PN 4304437 )
- 20x GE Sample Loading Reagent (Fluidigm, catalog number: PN 100-7610 )
- 2x Assay Loading Reagent (Fluidigm, catalog number: PN 100-7611 )
- 10x Assays for single-cell qPCR (see Recipes)
- Sample pre-mix (see Recipes)
Equipment
- Neubauer chamber
- Micropipettes (P2, P10, P20, P200, P1000)
- Multichannel pipette P10 with high precision (Rainin, catalog number: 17013802 )
- Cell culture incubator: 37 °C and 5% CO2 (Panasonic Healthcare, catalog number: MCO-170AICUVH-PJ )
- Clean bench (Panasonic, catalog number: MCV-131BNF-PJ )
- Centrifuge for 1.5 ml tubes (Eppendorf, model: 5418R )
- Centrifuge for 15 ml tubes (Tomy, model: Ax-511 )
- Centrifuge for 96-well plate (Sigma, model: 4-16KS )
- Vortex (Scientific Industries, model: Vortex-Genie 2 )
- Fluorescent confocal microscope (Keyence, model: BZ-X700 )
- Refrigerator and Freezer (any company’s product should be fine)
- C1 Auto Prep System (Fluidigm)
- IFC Controler HX (Fluidigm)
- BioMarkHD system for real-time qPCR (Fluidigm)
Software
- Fluidigm Real-Time PCR Analysis Software v.2.1.3
- Fluidigm Data Collection Software v.2.1.3
- Excel
- R (version 3.3.2)
Procedure
Single-cell qPCR protocol can be mainly divided into 4 fractions: The first one comprises cell culture and cell suspension preparation. The second and third are mainly done by the C1 integrated fluidic circuits, which allows single cell separation, cell lysis and pre-amplification. The final fraction is the single real time qPCR, which is done by the BioMarkHD system. Those steps are represented in Figure 1.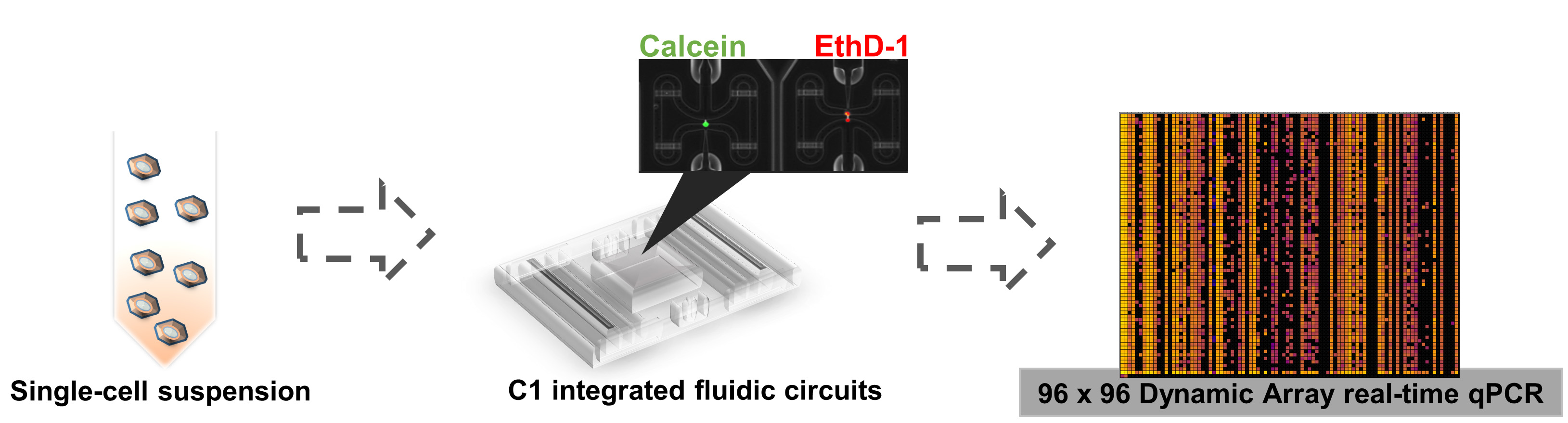
Figure 1. Scheme of single-cell qPCR protocol
- Sample preparation
This protocol commences with cell culture already ongoing.- When the cells reach 70% of confluence are ready to collect.
- Remove the medium and wash the dish with PBS once.
- Add 1 ml of TrypLE Express.
- Place the dishes in the 37 °C incubator, and incubate for 3-5 min until the cells detach.
- Add 4 ml of pre-warmed complete medium at 37 °C to stop the reaction.
- Pipette delicately to ensure the complete detach of all the cells but without damaging the cells.
- Transfer the cells into 15 ml tube.
- Centrifuge at 300 x g 4 °C for 5 min.
- Aspirate the medium and add 5 ml of PBS.
- Take apart 100 μl to count the cells.
- Centrifuge the 15 ml tube at 300 x g 4 °C for 5 min.
- While centrifuging, mix the separated cells at a proportion of 1:1 with Trypan blue solution.
- Insert 10 μl of the mixt into Neubauer chamber.
- Count the total cell number with the microscope (including dead cells).
- Remove PBS.
- Re-suspend the cells at 300 cells/μl (3 x 105 cells/ml) in Initial cell suspension buffer (Recipe 1) and keep on ice.
Note: An amount of at least 10,000 cells are recommended to obtain a complete capture, we observed that less than that amount leaves multiple channels with cells unattached in the IFC plate.
- Single-cell separation
- Take all the necessary reagents from -20 °C, where are stocked, and let them thaw on ice for 30 min before starting the experiment. Just before starting, take the reagents kept at 4 °C and put them on ice during its use.
Caution: The plate must not be placed on ice. - Remove the black plastic situated at the button of the integrated fluidic circuits (IFC) plate.
- Before starting, turn on the C1 machine since it takes some time to start up (about 3 min).
- Pipette several reagents following this scheme:
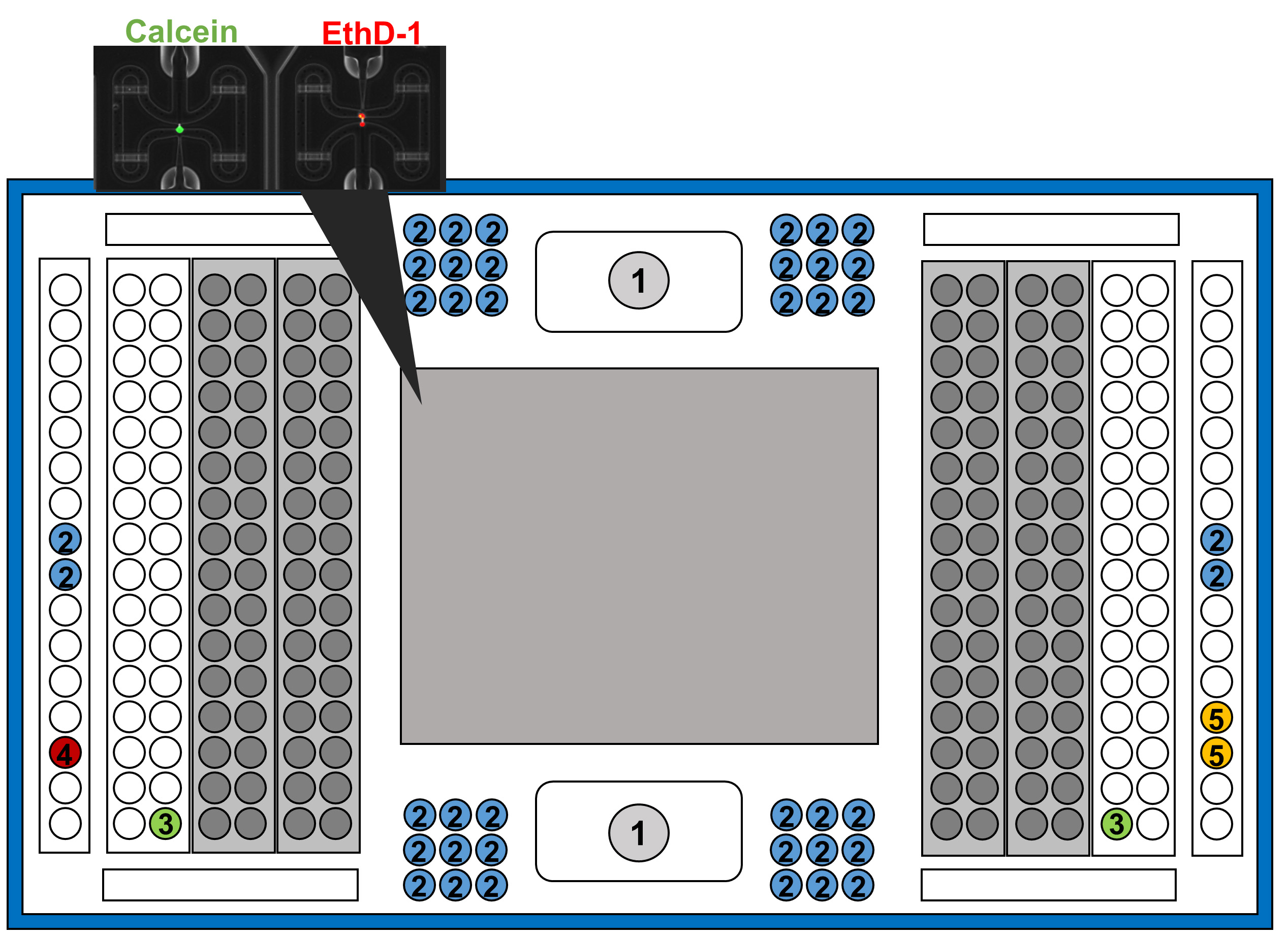
Figure 2. Representation of IFC plate indicating the wells to add the different reagents and solutions. Same number indicates same solution and volume. The wells covered in grey, represent the section of the chip that is covered by a film of paper, which at the end of the experiment will contain, each single-cell sample pre-amplified ready to use in sc-qPCR.
①.Add 200 μl of Harvest reagent. This well is like a button with a plastic lid on an iron spindle. To apply the reagent, press slightly with the tip, and once slips down, put the tip on the space. Then add slowly the reagent. As can be seen in Figure 3.
②.Add 20 μl of Harvest reagent.
③.Pipette 15 μl of Blocking reagent.
④.Add 20 μl of Preloading reagent.
⑤.Add 20 μl of Wash Buffer.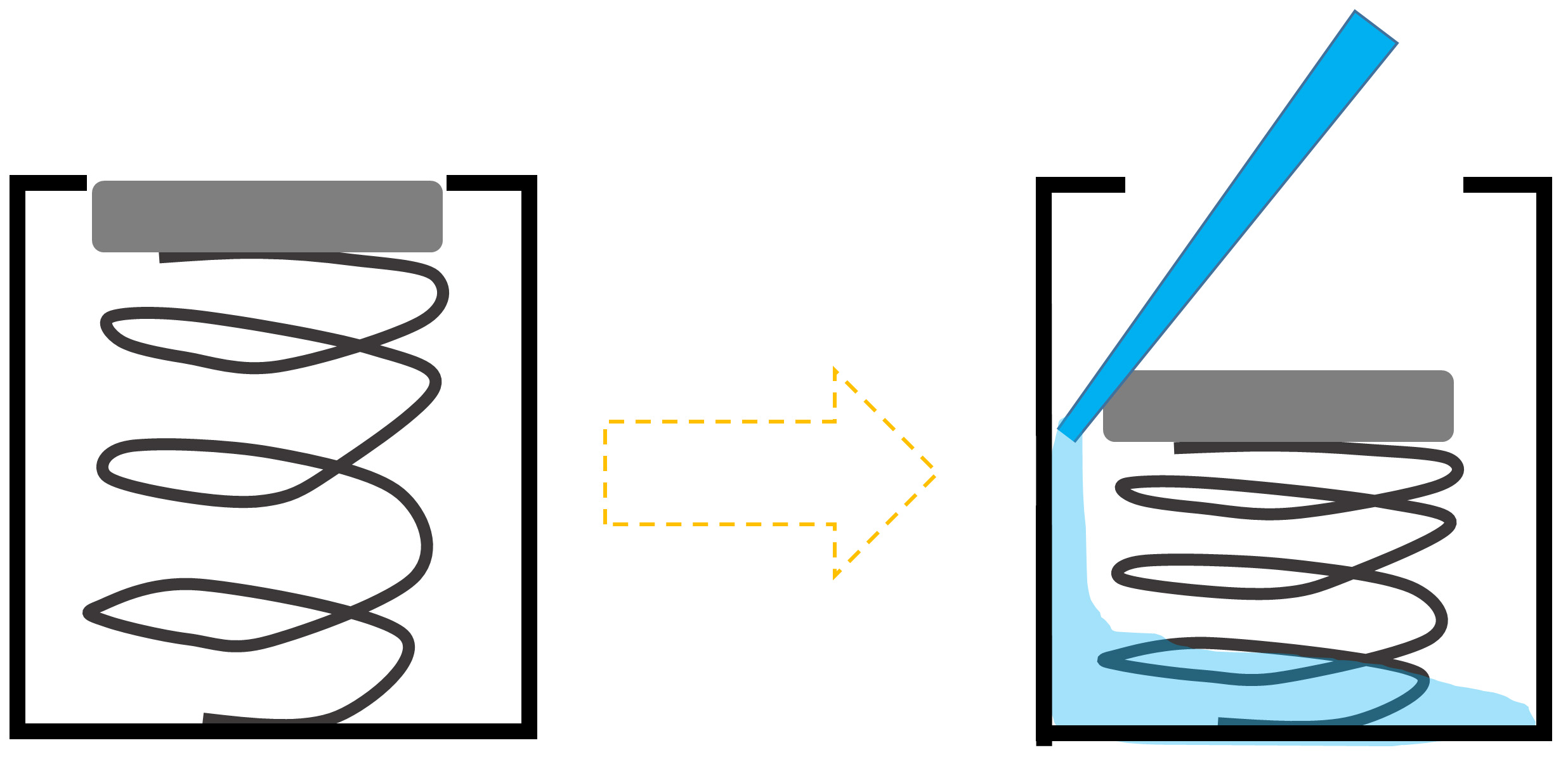
Figure 3. Scheme of Harvest reagent (200 μl) application in the IFC plate - Insert the plate in the IFC machine with the barcode facing outside.
Note: If the bottom of the plate is dirty, the machine will not detect it and will give an error. If so, wipe the bottom with ethanol 70% and place it again. - Run the mode STA:Prime (1782x/1783x/1784x) – according to the selected size of channels of the chip. This procedure takes 10-12 min.
While priming, prepare the cell mixture. - Prepare the cell mixture: 30 μl of cell suspension and 20 μl of suspension reagent.
Note: Less amount of cell mix can be prepared as long as the ratio is maintained. For instance, 6 μl and 4 μl. - After priming is finished, click eject and take out the plate.
- Remove the leftovers of C1 blocking reagent with a P200 pipette (wells are indicated in Figure 2-③).
- Insert the cells and Live/Dead staining solutions, as follows:
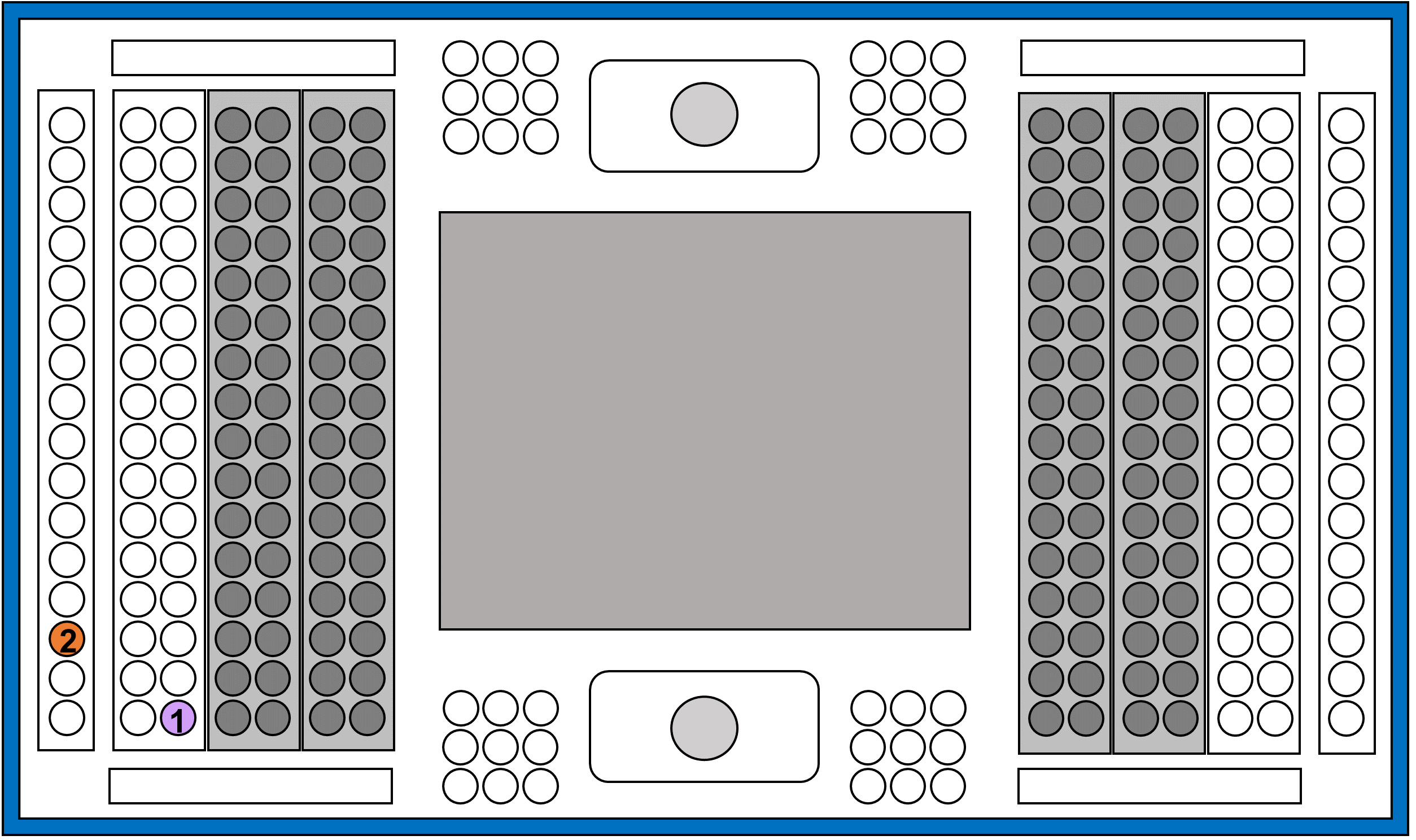
Figure 4. Representation of IFC plate indicating the wells to add the different reagents and solutions
①.Add 6 μl of cell mixture.
Note: Do not vortex the cells. Pipette them before the addition.
②.Add 20 μl of Live/Dead staining solution (Recipe 2). - Place the IFC plate into the C1 system.
- Select STA: Cell Load&Stain (1782x/1783x/1784x)–according to the selected size of channels of the chip. It takes 30-65 min, depending on the cell size of the plate.
- Meanwhile, prepare the Lysis Mix, RT Mix and PreAmp Mix (Recipes 3, 4 and 5).
- When Loading is complete, withdraw the plate from the C1 and take pictures of the cells with Keyence microscope. Using the Phase contrast at 10x, channels and fluorescence-stained cells can be easily observed on the central part of the plate, colored in grey color in Figure 4. Also each well number can also be observed.
- Write down the channels with dead cells, empty channels and channels with more than one cell to be discarded a posteriori (Figure 5).
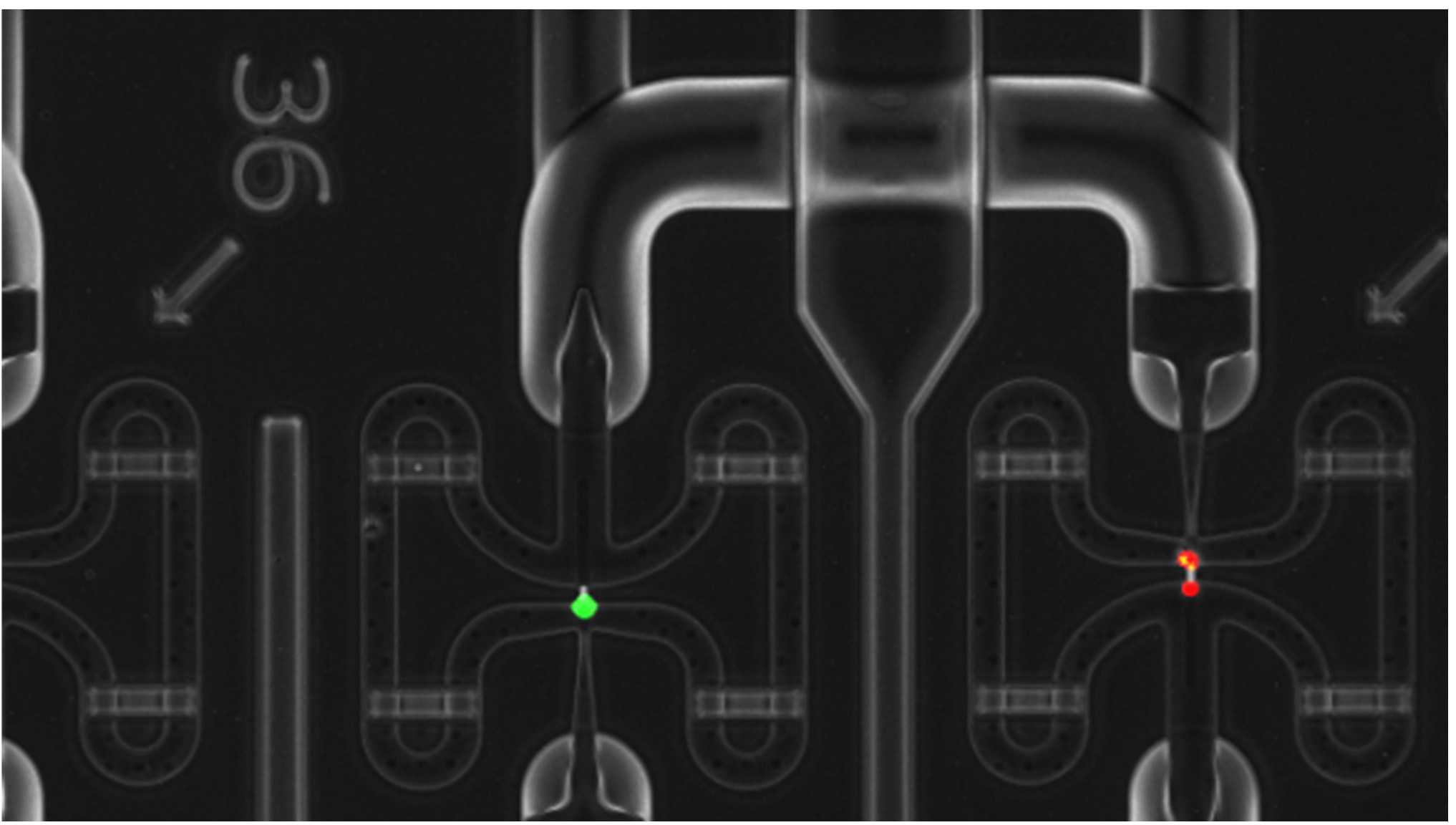
Figure 5. Example of Live/Dead staining. Green cells are living cells while read are dead, or highly damaged cells. Moreover, right chamber shows a doublet, and left one shows a clear single cell.
- Take all the necessary reagents from -20 °C, where are stocked, and let them thaw on ice for 30 min before starting the experiment. Just before starting, take the reagents kept at 4 °C and put them on ice during its use.
- Cell lysis, reverse transcription and pre-amplification.
- Add the reagents following the scheme in Figure 6:
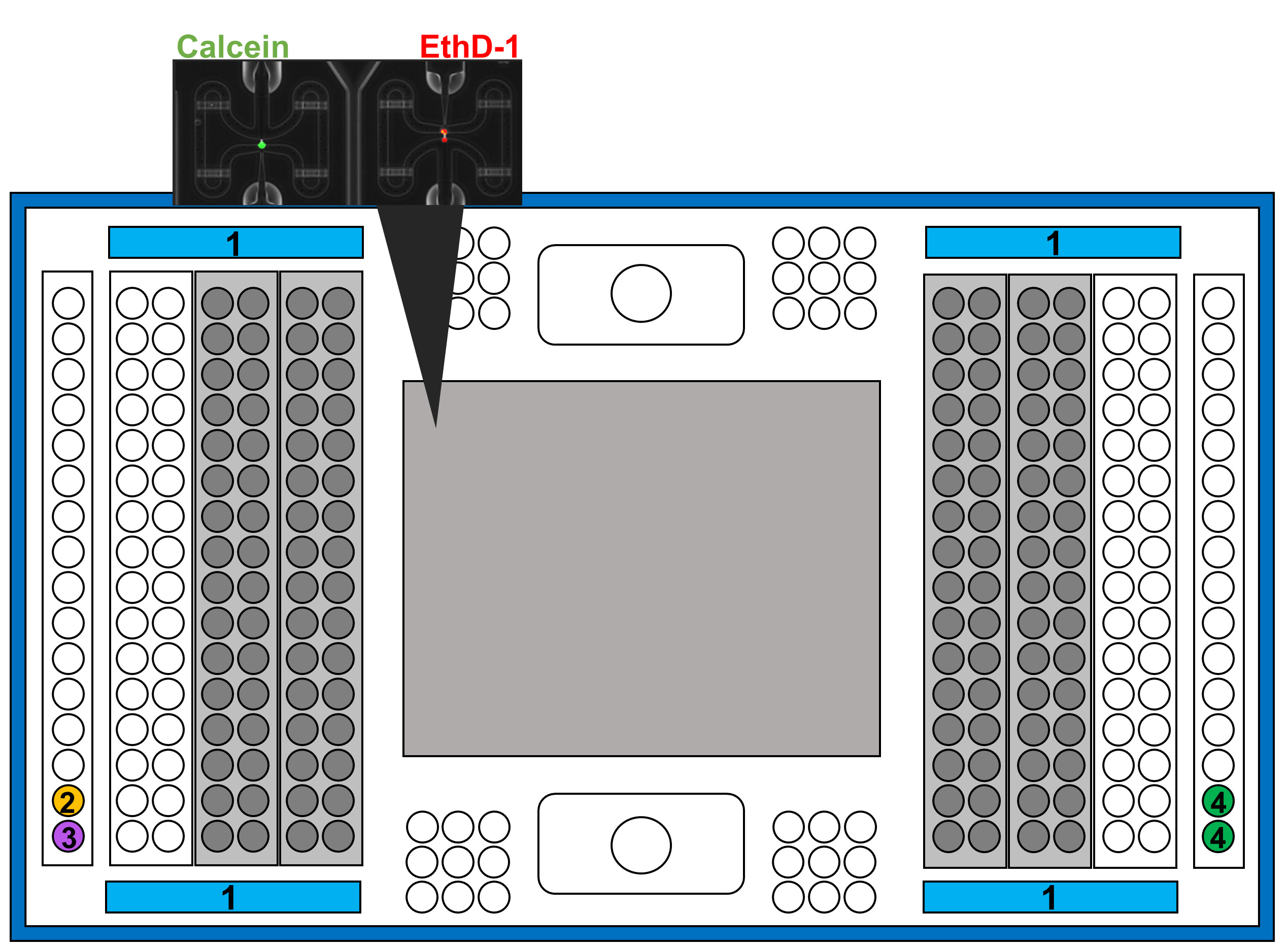
Figure 6. Representation of IFC plate indicating the wells to add the different reagents and solutions
①.Add 180 μl of Harvest reagent. Carefully move the plate from side-to-side to ensure that the reagent covers all the surface.
②.Add 7 μl of Lysis Final Fix.
③.Add 7 μl of RT Final mix.
④.Add 24 μl of Pre-amplification Mix (Recipe 6). - Place the plate into the C1 system.
- Select STA: PreAmp (1782x/1783x/1784x)–according to the selected size of channels of the chip- and choose the finish hour. We usually leave it overnight and collect it next morning to proceed with the experiments. The process itself takes 5-6 h approximately.
- When STA: Preamp process has finished, select EJECT and remove the IFC plate.
- Using a new PCR 96-well plate, add 25 μl of C1 dilution reagent in each well.
- Remove the papers than protect the output wells on IFC plate (those covers are represented in grey color in Figures 2, 4 and 6).
- Using an eight-channel micropipette (Rainin), transfer the samples onto the 96-well plate.
Note: The amount of sample should be close to 3 μl, but to ensure, please set the micropipette at 4 μl. - Transfer the samples using this specific order, described in Figure 7.
Note: Using 8-channel micropipette, and taking in consideration that IFC plate has 16 well per lane, it has the specific size to pic up every-other well.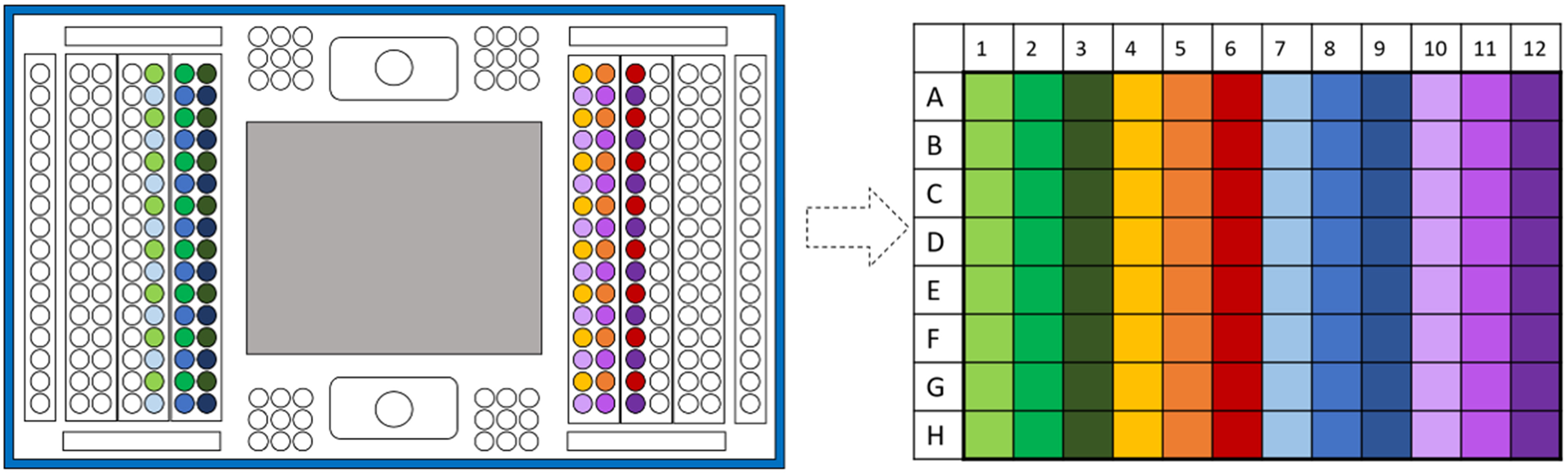
Figure 7. Scheme to illustrate the order of well transfer from IFC plate to PCR 96-well plate - Vortex the 96-well plate and spin down. At this point, the samples can be frozen at -20 °C and be kept for few weeks before use, in that case, seal the plate with a PCR-plate film.
- Add the reagents following the scheme in Figure 6:
- Single-cell qPCR
- Take all the necessary reagents from -20 °C, where are stocked, and let them thaw on ice for 30 min before starting the experiment. Just before starting, take the reagents kept at 4 °C and put them on ice during its use.
Caution: The plate must not be placed on ice. - Turn on the Standard 96-well Thermal Cycler, because it takes some time to start up.
- Remove the blue protective film from the bottom of the 96.96 chip.
- Using the syringe from the kit, inject the control line fluid in the two big central wells, pressing between the cross (The cross can be seen, marked with an arrow in Figure 8).
Note: The oil leaks easily, so be careful when managing the syringe.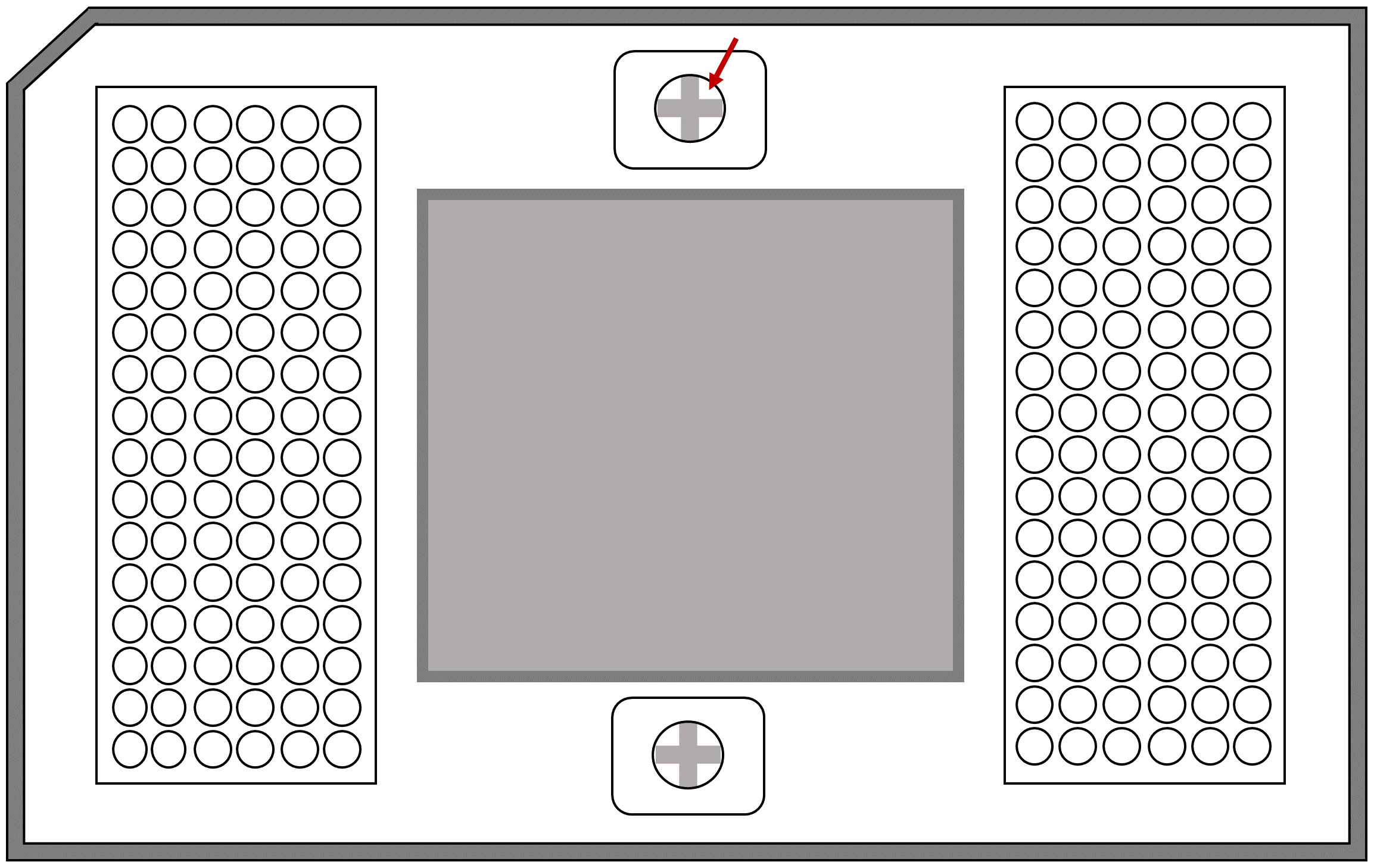
Figure 8. Schematic representation of the 96.96 quantitative PCR Dynamic Array microfluidic chip. The arrow shows the space in where the line fluid should be applied. - Insert the plate (with the barcode facing the outside).
- Select the option Prime(136x). This process takes approximately 20 min.
- Meanwhile, prepare the Pre-mix sample (Recipe 8).
- In a new PCR 96-well plate, add 3.3 μl of sample pre-mix in each well.
- Add 2.7 μl of cDNA sample with a multichannel micropipette (Sample obtained at Step C9) making a total volume of 6 μl in each inlet.
- Seal with PCR-plate film.
- Prepare the Taqman assays Mix (Recipe 7) and seal the plate with PCR-plate film.
- Vortex both plates for 10 sec and spin down.
- When priming is complete, collect the chip.
- Transfer the samples and the Taqman assays. In the following order (Figures 9 and 10):
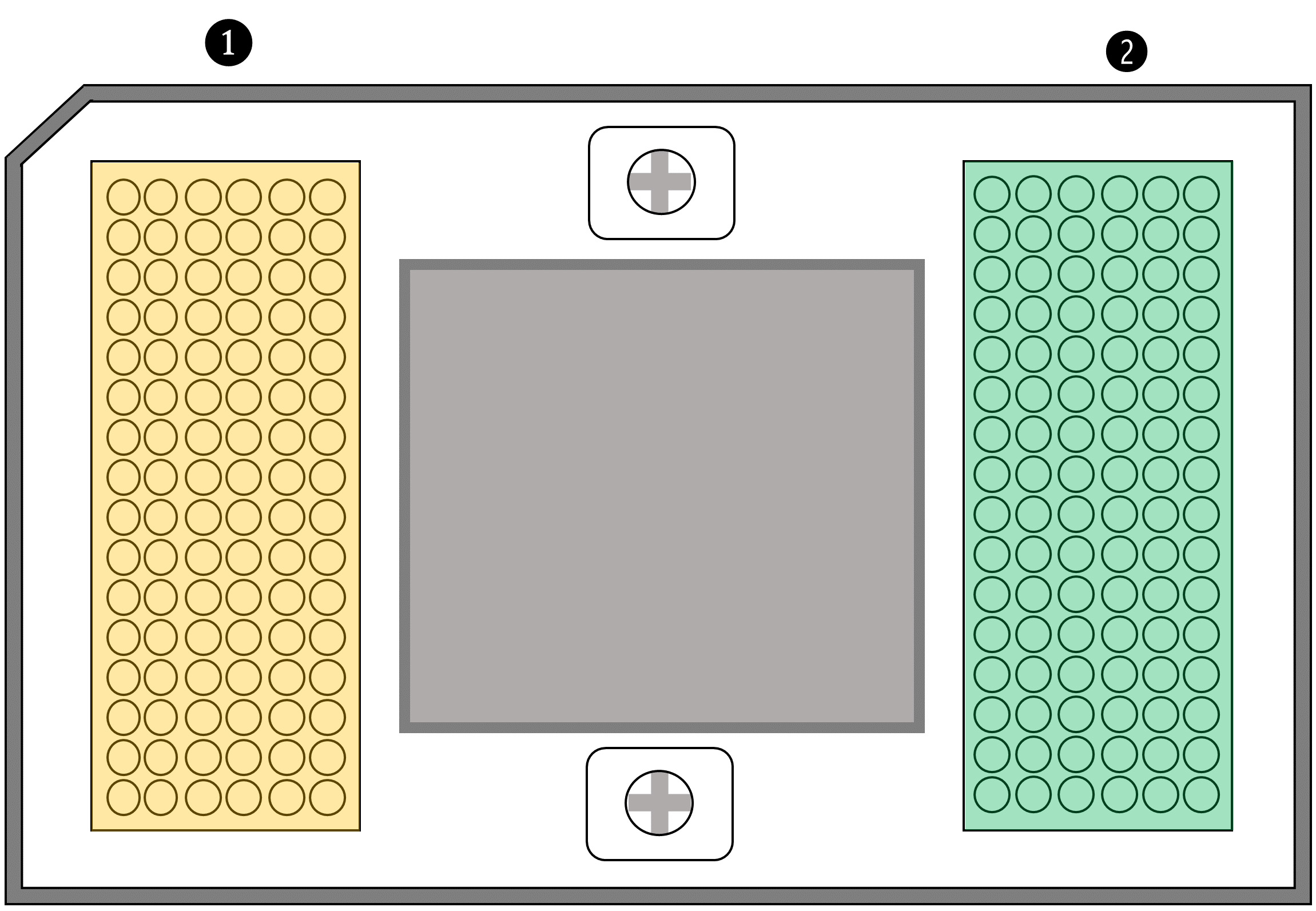
Figure 9. Scheme of 96.96 chip showing in yellow color the inlets to insert Taqman assays and, in green, the inlets to insert sample premix
①.Add 5 μl of Taqman assays in the yellow inlets, using the following order (Figure 10A) to transfer from qPCR 96-well plate to IFC chip.
②.Add 5 μl of Sample Premix, using the same order (Figure 10A) in the green inlets.
Notes:- The inlet wall is not horizontal, but has a diagonal angle. To ensure better application; the best option is insert the tip inclined, as is illustrated in Figure 10B.
- Avoid bubbles when pipetting. However, if there are bubbles but they move, means that they are on the surface, and then they will not interfere in the analysis. If bubbles do not move, with a micropipette tip remove the bubble.
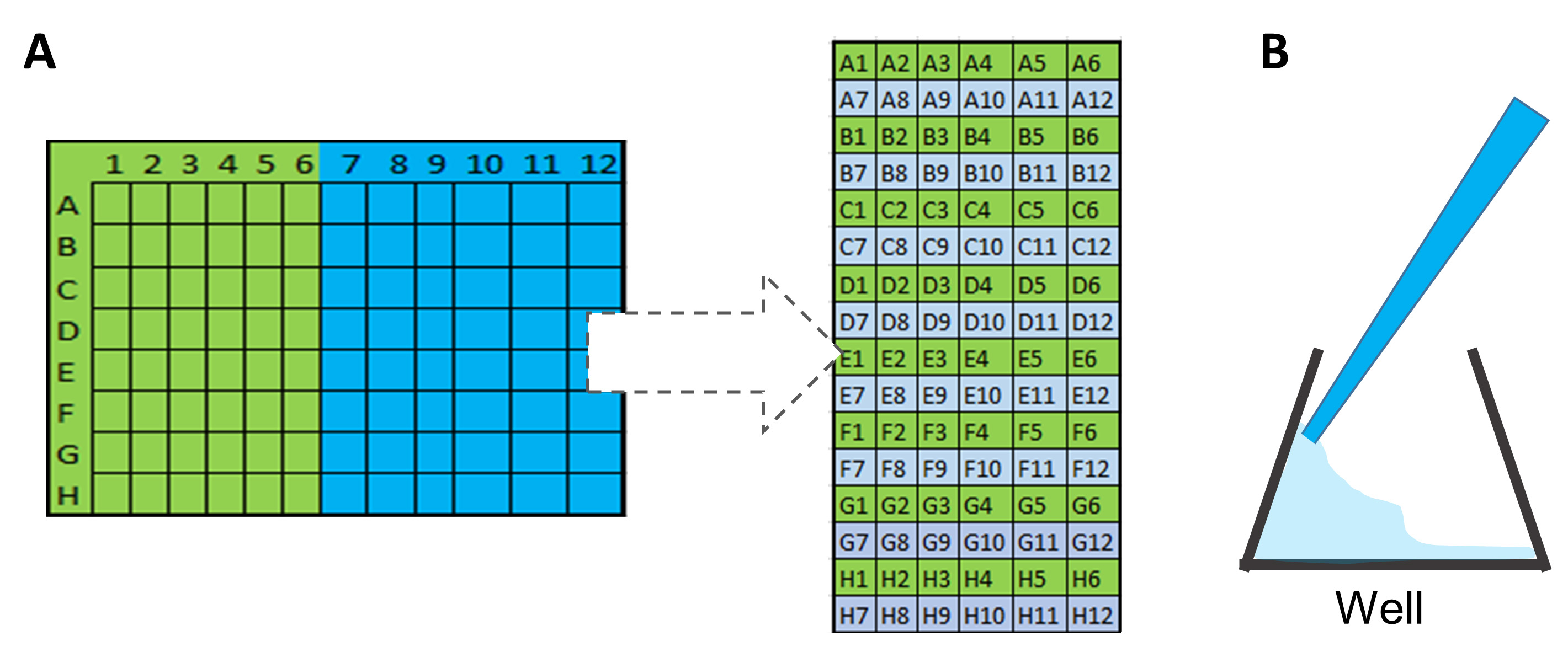
Figure 10. Scheme of order and methodology to transfer samples into 96.96 chip. A. Scheme to illustrate the order of well transfer from PCR plate to 96.96 chip. B. Scheme of sample application in 96.96 inlets. - Insert the chip into the IFC Controller HX and run the script Load Mix (136x).
- While running the Load script, turn on the computer connected to the BioMarkHD system for real-time qPCR.
- Run the software Biomark data collection.
Double click to turn on the option “lamp” to start up. This process takes approximately 20-30 min. - When the Loading finish, bring the chip to BioMarkHD system and follow these steps:
- Click “Start a new run”.
- Insert the chip with the bar code facing outside.
Select: Load
→Next - Select the settings of the 96.96 chip.
- Choose the name and the folder where you want to save the document.
→Next - Application type: Single probe.
- Probes: FAM_MGB
→Next - Protocol: GE 96x96 Standard v1.
→Next
- Choose the name and the folder where you want to save the document.
- Start the run.
- Once it finishes remove the plate, close the software and shut down the computer.
- Take all the necessary reagents from -20 °C, where are stocked, and let them thaw on ice for 30 min before starting the experiment. Just before starting, take the reagents kept at 4 °C and put them on ice during its use.
Data analysis
The data analysis procedure refers to our previous report (Prieto-Vila et al., 2019).
- Remove dead/damaged cells
- After running the qPCR, collect the raw data to be analyzed with the software Biomark.
- Use the settings Linear (derivative) and User (detectors) settings to generate Ct values for each gene.
- Remove all values with confidence lower than 0.2.
- Export the data to excel file.
- Using the data obtained in Step B14 from the protocol (pictures of Live/Dead staining), remove all the cells that were stained red (dead), chambers with more than one cell, or none.
- Remove cells whose housekeeping genes have Ct values lower than 16. In our case, we used both BACTIN and GAPDH.
- Data normalization
Normalization can be conducted by two systems:- With the objective to remove the variation between assays, normalization can be done using a housekeeping gene; in our case, we utilized GAPDH. The normalization was calculated by the formula: 2-ΔΔCT.
- Another broadly used methodology in single-cell analysis is the normalization by log2Ex, which expresses the transcript levels above the background in log base 2 (Livak et al., 2013). In our case, we set the LOD (limit of detection) to 26, as it was the average of the lowest detectable Ct value for each gene. This can calculated by the formula: Log2Ex = LOD (Limit Of Detection) Cq – Cq [Gene].
- Both normalizations can be done with Excel.
- For a more detailed protocol containing examples, please refer to this manual: “Singular: Analysis Toolset” (P/N 100-5066) https://www.fluidigm.com/binaries/content/assets/fluidigm/singular-analysis-toolset-3.5/singular-analysis-toolset-3.5-userguide.pdf.
- Several statistical analysis can be done for single-cell qPCR
Probably the most common statistical analysis for single-cell are t-SNE Plot, Heatmap and ViolinPlots.
For Heatmap we used the software Partek Genomics Suite Software; while both t-SNE and Violin Plot were done by the free software R using the package “ggplot2”.
Notes
- For single-cell analyses, it is considered that rather than do replicates, which is also physically impossible, is more important to increase the sample number. Thus, we ran each sample twice. Analyzing a total of 192 individual cells per sample.
- There are three sizes of C1 IFC plates to adjust the channel to the different sizes of cells. Prior to choose the size of the plate, we calculated the cell size by taking pictures of the cells after being detached, and measuring the size by Keyence software. The available sizes are: S (5-10 μm), M (10-17 μm), L (17-25 μm). As a reference, we used M size for MDA-MM-231 and MCF7 cells; and we have run hepatocytes in L size plate (Katsuda et al., 2019).
- One of the most important steps is single-cell isolation. During this process, cells are isolated from their local environment, and thus, keeping those cells alive and in good condition for proper further analyses is indispensable (Van Den Brink et al., 2017). Please manage them carefully.
- This protocol has been done following the Fluidigm system manufacture’s protocols (PN-100-6177 J1 and PN 68000130 E1), which can be found at: https://www.fluidigm.com/binaries/content/documents/fluidigm/resources/c1-taqman-pr-100%E2%80%906117/c1-taqman-pr-100%E2%80%906117/fluidigm%3Afile and https://www.fluidigm.com/binaries/content/documents/fluidigm/resources/96.96-ge-taqman%E2%80%90std-qr-68000130/96.96-ge-taqman%E2%80%90std-qr-68000130/fluidigm%3Afile respectively.
Recipes
Note: All the recipes, unless otherwise stated, are the reagent necessary for one run.
- Initial cell suspension buffer
ComponentsVolume
*This solution can be used multiple times and should be stored at 4 °C.Components Volume EDTA (5M) 0.1 ml FBS 50 μl PBS 10 ml - LIVE/DEAD Cell Staining solution (Total 1253.13 μl)
Components Volume Cell Wash Buffer (C1 Single-Cell Auto Prep Reagent Kit) 1,250 μl Ethidium homodimer-1 (Live/Dead kit) 2.5 μl Calcein AM (Live/Dead kit) 0.63 μl - Lysis Mix (Total 18.1 μl)
*Vortex thoroughlyComponents Volume Single Cell Lysis Solution (Ambion single cell-to-Ct qRT-PCR Kit) 12.75 μl C1 Lysis Plus Reagent (C1 Single-Cell Auto Prep Reagent Kit) 4.35 μl C1 DNA Dilution Reagent (C1 Single-Cell Auto Prep Reagent Kit) 1 μl - Reverse transcription Final Mix (Total 12.4 μl)
*Vortex thoroughlyComponents Volume Stop Solution (Ambion single cell-to-Ct qRT-PCR Kit) 1.94 μl Single cell VILO RT Mix (Ambion single cell-to-Ct qRT-PCR Kit) 5.84 μl Single Cell SuperScript RT (Ambion single cell-to-Ct qRT-PCR Kit) 3.62 μl Loading reagent (C1 Single-Cell Auto Prep Reagent Kit) 1 μl - 0.2x Pooled Taqman Primers (Total 100 μl)
*Vortex thoroughlyComponents Volume 1 μl of each Taqman Probe (x96 probes) 1 μl each C1 DNA Dilution Reagent (C1 Single-Cell Auto Prep Reagent Kit) 4 μl
**This reagent can be made in advance and aliquots stocked up to 6 months at -20 °C. - Pre-amplification Mix (Total 60 μM)
*Vortex thoroughlyComponents Volume Single-Cell PreAm Mix (Ambion single cell-to-Ct qRT-PCR Kit) 12 μl Loading Reagent (C1 Single-Cell Auto Prep Reagent Kit) 3 μl Pooled primers (The mix previously prepared) 15 μl DNA-free water 30 μl - 10x Assays for single-cell qPCR
In a qPCR 96-well plate, in each inlet add the following mixture for each of the Taqman assays.
**This reagent can be made in advance and stocked up to 6 months at -20 °CComponents Volume per inlet Volume 10 reactions stock 20x Taqman gene expression Assay 2.5 μl 25 μl 2x Assay Loading Reagent 2.5 μl 25 μl - Sample pre-mix
Components Volume per inlet Sample pre-mix for all wells Taqman Universal PCR Master Mix 2.5 μl 360 μl 20X GE Sample Loading Reagent 0.25 μl 36 μl
Acknowledgments
The present work was supported in part by a Grant-in-Aid for Scientific Research (C) JSPS KAKENHI Grant Number: 19K16761, Grant-in-Aid for Young Scientists (A) JSPS KAKENHI Grant Number: 17H04991 and the “Development of Diagnostic Technology for Detection of miRNA in Body Fluids” grant from the Japan Agency for Medical Research and Development (AMED).
This protocol was adapted from Fluidigm system manufacture’s protocols (PN-100-6177 J1 and PN 68000130 E1).
Competing interests
We have no conflict of interest to declare.
References
- Karaiskos, N., Wahle, P., Alles, J., Boltengagen, A., Ayoub, S., Kipar, C., Kocks, C., Rajewsky, N. and Zinzen, R. P. (2017). The Drosophila embryo at single-cell transcriptome resolution. Science 358(6360): 194-199.
- Katsuda, T., Hosaka, K., Matsuzaki, J., Usuba, W., Prieto-Vila, M., Yamaguchi, T., Tsuchiya, A., Terai, S. and Ochiya, T. (2019). Transcriptomic dissection of hepatocyte heterogeneity: linking ploidy, zonation, and stem/progenitor cell characteristics. Cell Mol Gastroenterol Hepatol.
- Livak, K. J., Wills, Q. F., Tipping, A. J., Datta, K., Mittal, R., Goldson, A. J., Sexton, D. W. and Holmes, C. C. (2013). Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods 59(1): 71-79.
- Prieto-Vila, M., Usuba, W., Takahashi, R. U., Shimomura, I., Sasaki, H., Ochiya, T. and Yamamoto, Y. (2019). Single-cell analysis reveals a preexisting drug-resistant subpopulation in the luminal breast cancer subtype. Cancer Res 79(17): 4412-4425.
- Prieto-vila, M., Yamamoto, Y., Takahashi, R. Ochiya, T. (2018). Single-cell transcriptomics. In: Handbook of Single Cell Technologies. Springer, Singapore.
- Rato, S., Golumbeanu, M., Telenti, A. and Ciuffi, A. (2017). Exploring viral infection using single-cell sequencing. Virus Res 239: 55-68.
- Van den Brink, S. C., Sage, F., Vértesy, Á., Spanjaard, B., Peterson-Maduro, J., Baron, C. S., Robin, C. and van Oudenaarden, A. (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods 14(10): 935-936.
- Wen, L. and Tang, F. (2016). Single-cell sequencing in stem cell biology. Genome Biol 17: 71.
- Woyke, T., Doud, D. F. R. and Schulz, F. (2017). The trajectory of microbial single-cell sequencing. Nat Methods 14(11): 1045-1054.
- Zheng, C., Zheng, L., Yoo, J. K., Guo, H., Zhang, Y., Guo, X., Kang, B., Hu, R., Huang, J. Y., Zhang, Q., Liu, Z., Dong, M., Hu, X., Ouyang, W., Peng, J. and Zhang, Z. (2017). Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169(7): 1342-1356 e1316.
- Ziegenhain, C., Vieth, B., Parekh, S., Reinius, B., Guillaumet-Adkins, A., Smets, M., Leonhardt, H., Heyn, H., Hellmann, I. and Enard, W. (2017). Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 65(4): 631-643 e634.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Prieto-Vila, M., Ochiya, T. and Yamamoto, Y. (2020). Single-cell qPCR Assay with Massively Parallel Microfluidic System. Bio-protocol 10(6): e3563. DOI: 10.21769/BioProtoc.3563.
Category
Cancer Biology > General technique > Genetics > Gene expression
Biochemistry > RNA > Single-molecule Activity
Molecular Biology > RNA > qRT-PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









