- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mesenchymal Stromal Cells Derived from Bone Marrow and Adipose Tissue: Isolation, Culture, Characterization and Differentiation
Published: Vol 10, Iss 4, Feb 20, 2020 DOI: 10.21769/BioProtoc.3534 Views: 8853
Reviewed by: Ralph Thomas BoettcherShweta SharmaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2411 Views

Reprogramming White Fat Cells for Adipose Manipulation Transplantation (AMT) Therapy
Kelly An [...] Nadav Ahituv
Aug 5, 2025 2240 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 211 Views
Abstract
Since their discovery, mesenchymal stromal cells (MSCs) have received a lot of attention, mainly due to their self-renewal potential and multilineage differentiation capacity. For these reasons, MSCs are a useful tool in cell biology and regenerative medicine. In this article, we describe protocols to isolate MSCs from bone marrow (BM-MSCs) and adipose tissues (AT-MSCs), and methods to culture, characterize, and differentiate MSCs into osteoblasts, adipocytes, and chondrocytes. After the harvesting of cells from bone marrow by flushing the femoral diaphysis and enzymatic digestion of abdominal and inguinal adipose tissues, MSCs are selected by their adherence to the plastic tissue culture dish. Within 7 days, MSCs reach 70% confluence and are ready to be used in subsequent experiments. The protocols described here are easy to perform, cost-efficient, require minimal time, and yield a cell population rich in MSCs.
Keywords: Adipose tissueBackground
The concept of stem cells dates back to the 19th century, but their existence was confirmed in the 1960s and 1970s following experiments by Friedenstein and collaborators, which showed the presence of stem cells in the bone marrow (Friedenstein, 1970; Bianco et al., 2008). Afterward, Caplan (1991) named them as mesenchymal stem cells (here, called mesenchymal stromal cells–MSCs) and proposed their use in regenerative medicine. In the bone marrow, the percentage of MSCc is estimated to be 0.001 to 0.01% of the total mononuclear cells. Because of their scarcity, alternative sources have been described, although bone marrow remains as the main source of MSCs (Nancarrow-Lei et al., 2017). Adipose tissue is a very promising source because it contains a large number of MSCs that are relatively easy to harvest with minimal discomfort and risk for donors (Zuk et al., 2001). The protocols used to harvest and culture MSCs from either bone marrow (BM-MSCs) or adipose tissue (AT-MSCs), may vary among different species or even among different strains of the same species. The most commonly used methods for obtaining MCSs involve using flow cytometry (Schrepfer et al., 2007), multipotent adult progenitor cell media (Harting et al., 2008), the ficoll-paque gradient centrifugation method (Pierini et al., 2012), and immunomagnetic beads (Wadajkar et al., 2014). Here, we describe cost-efficient protocols that are relatively easy and fast to perform and can be used to obtain cell populations rich in MSCs from bone marrow and adipose tissues. These protocols can be used to study several cellular and molecular aspects of MSCs, such as their proliferation, differentiation, and signaling pathways (Abuna et al., 2016; Fideles et al., 2019), the biological effects of growth factors and drugs on MSCs (Oliveira et al., 2012; Zhang et al., 2017), the interactions between MSCs and natural or synthetic biomaterials (Hu et al., 2018; Lopes et al., 2019), and the application of MSCs in regenerative medicine strategies (Almeida et al., 2019; Freitas et al., 2019).
Materials and Reagents
- Sterile surgical drape
- Aluminum foil
- Coat (ProtDesc, catalog number: 80404440020), storage temperature: RT
- Mask (ProtDesc, catalog number: 80404440006), storage temperature: RT
- Cap (ProtDesc, catalog number: 80404440004), storage temperature: RT
- Gloves (Maxitec, Kevenol, catalog number: 80748910002), storage temperature: RT
- 20-ml syringe (BD Plastipak, catalog number: 990687), storage temperature: RT
- 21G needle (BD PrecisionGlide, catalog number: 300054), storage temperature: RT
- Glass tissue culture dish (Pyrex, catalog number: HX0004-00376), storage temperature: RT
- Corning® 75 cm2, U-Shaped canted neck cell culture flask with vent cap (Corning, catalog number: 430641U), storage temperature: 15/30 °C
- 24-well cell culture plates (Corning, catalog number: 3524), storage temperature: 15/30 °C
- 12-well plates (Corning, catalog number: 3512), storage temperature: 15/30 °C
- 6-well culture plates (Corning, catalog number: 3335), storage temperature: 15/30 °C
- 50-ml conical tube (Sarstedt, catalog number: 62.547.254), storage temperature: 15/30 °C
- Microtube 1.5-ml (Eppendorf, catalog number: Z606340), storage temperature: RT
- Micropipette tips (Eppendorf, catalog numbers: 0030000811/0030000854/0030000870/0030000919), storage temperature: RT
- Ultra-low attachment, 96-well (Costar, catalog number: CLS7007), storage temperature: 15/30 °C
- Alpha minimum essential medium (α-MEM) (Thermo Fisher Scientific, catalog number: 12000-022), storage temperature: 2/8 °C
- Dulbecco’s modified Eagle’s medium (D-MEM) (Thermo Fisher Scientific, catalog number: 12100-046), storage temperature: 2/8 °C
- Dulbecco’s phosphate-buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 21600-010), storage temperature: 15/30 °C
- Sodium bicarbonate (Sigma-Aldrich, Sigma, catalog number: S5761-1KG), storage temperature: 15/30 °C
- Gentamycin reagent solution (Thermo Fisher Scientific, catalog number: 15710-064), storage temperature: -20/-5 °C
- Penicillin-Streptomycin (Thermo Fisher Scientific, catalog number: 15140-122), storage temperature: 15/30 °C
- Dexamethasone (Sigma-Aldrich, catalog number: D8893), storage temperature: 2/8 °C
- FBS qualified fetal calf serum (Thermo Fisher Scientific, catalog number: 12657-029), storage temperature: -10 °C
- Amphotericin B 250 μg/ml (Thermo Fisher Scientific, catalog number: 15290-018, storage temperature: -20/-5 °C)
- 0.25% Trypsin (1x) (Thermo Fisher Scientific, catalog number: 15050-057), storage temperature: -20/-5 °C
- Collagenase type II lyophilized (Thermo Fisher Scientific, catalog number: 17101-015), storage temperature: 2/8 °C
- 2.5% Chlorhexidine (Bioflora Manipullarium), storage temperature: RT
- β-Glycerophosphate disodium salt pentahydrate 98.0% (NT) (Sigma-Aldrich, catalog number: 50020-100G), storage temperature: 2/8 °C
- L-Ascorbic acid (Sigma-Aldrich, catalog number: 33034-100G), storage temperature: 15/30 °C
- Ethanol 96% (Merck, catalog number: 100971), storage temperature: 5/30 °C
- Formaldehyde solution 37% (Merck, catalog number: 104002), storage temperature: 15/25 °C
- Isopropanol (Merck Millipore, catalog number:1096341000), storage temperature: 5/30 °C
- Alizarin red S (Sigma-Aldrich, catalog number: A5533-25G), storage temperature: 15/30 °C
- Acetic acid (Merck, catalog number: 199061), storage temperature: 15/25 °C
- 3-Isobutyl-1-methylxanthine (Sigma-Aldrich, catalog number: I7018-1000MG), storage temperature: -20 °C
- Methanol (Merck, catalog number: 1.06009), storage temperature: 5/30 °C
- Insulin human (Sigma-Aldrich, catalog number: I2643-50MG), storage temperature: -20 °C
- Hydrochloric acid fuming 37% (Merck, catalog number: 1.00317), storage temperature: 5/30 °C
- Indomethacin (Sigma-Aldrich, catalog number: I7378-5G, storage temperature: 15/30 °C)
- Oil red O (Sigma-Aldrich, catalog number: O0625-25G, storage temperature: 15/30 °C)
- Trichome stain (Masson) Kit (Sigma-Aldrich, catalog number: HT15-1KT, storage temperature: RT)
- Sodium pyruvate (Sigma-Aldrich, catalog number: S8636, storage temperature: 2/8 °C)
- Human albumin (Institute Grifols, catalog number: A4AFC03441, storage temperature: 2/25 °C)
- Transforming growth factor-β3 (Peprotech Inc., catalog number: 100-36E, storage temperature: -20 °C)
- 4% Paraformaldehyde (Electron Microscopy Sciences, catalog number: 157-4-100), storage temperature: 2/8 °C
- Xylene (LabSynth, catalog number: X1001.01.BJ), storage temperature: 16/26 °C
- Paraffin (EasyPath, catalog number: EP-21-20068A), storage temperature: 15/30 °C
- Eosin (Sigma-Aldrich, catalog number: HT110132), storage temperature: RT
- Monoclonal anti-rat antibody: anti-CD29 (BD Biosciences, catalog number: 562154, storage temperature: 4 °C)
- Monoclonal anti-rat antibody: anti-CD31 (BD Biosciences, catalog number: 555027, storage temperature: 4 °C)
- Monoclonal anti-rat antibody: anti-CD34 (Invitrogen, catalog number: 11-0341-81, storage temperature: 4 °C)
- Monoclonal anti-rat antibody: anti-CD45 (BD Biosciences, catalog number: 554878, storage temperature: 4 °C)
- Monoclonal anti-rat antibody: anti-CD90 (BD Biosciences, catalog number: 554898, storage temperature: 4 °C)
- Monoclonal anti-rat antibody: anti-CD106 (BD Biosciences, catalog number: 559229, storage temperature: 4 °C)
- Transport medium (see Recipes)
- Collagenase solution (see Recipes)
- Trypsin solution (see Recipes)
- Ascorbic acid and β-Glycerophosphate solution (see Recipes)
- Growth medium (10% MEM) (see Recipes)
- Osteogenic differentiation medium (see Recipes)
- Chondrogenic differentiation medium (see Recipes)
- Adipocyte differentiation medium (see Recipes)
- Dexamethasone stock solution (200 μM) (see Recipes)
- Ascorbic acid stock solution (20 mM) (see Recipes)
- TGF-β3 (see Recipes)
- Oil red O staining (see Recipes)
Equipment
- Scissors (Quinelato, catalog number: QT.109.14)
- Forceps (Quinelato, catalog number: QC.301.14)
- Erv-Mount® (EasyPath, catalog number: EP-51-05041), storage temperature: 20 °C
- Micropipette (Eppendorf, catalog numbers: 4921000028/4921000044/4921000079/4921000109/4921000117/4921000150)
- Analytical balance M214A (BEL, catalog number: BL0003)
- RT basic series magnetic stirrers (Thermo Fisher Scientific, catalog number: 88880009)
- Bench meter for pH (Hanna, catalog number: HI5522-01)
- Stericup quick release vacuum driven disposable filtration system (Merck, catalog number: S2GPU05RE)
- Vacuum pump and compressor (Prismatec, catalog number: 132)
- Airstream class II biohazard safety cabinet (Esco Micro Pte.Ltd., model: AC2-4E8)
- Microprocessor water bath (Quimis, catalog number: Q215M)
- CO2 incubator (Panasonic, Panasonic/Sanyo, model: MCO-19AIC)
- Eppendorf® Centrifuge 5702 (Sigma-Aldrich, catalog number: Z606936)
- Axiovert 25 inverted microscope for advanced routine (Carl Zeiss)
- Compact digital microplate shaker (Thermo Fisher Scientific, catalog number: 88880023)
- Epoch 2 microplate spectrophotometer (BioTek, catalog number: BTEPOCH2)
- Centrifuge 5418 R (Eppendorf, catalog number: 5401000013)
- Gas exhaust chapel (Lutech, catalog number: LCE-15)
- Vertical freezer, 231 liters (Consul, catalog number: CVU26EB)
- Refrigerator frost free, 342 liters (Consul, catalog number: CRB39AB)
- Ultra-low freezer (Panasonic, catalog number: MDF-U500VXC-PA)
- FACSCantoTM II (BD Biosciences, catalog number: 338962)
- Paraffin dispenser (Oma, catalog number: IO-88)
- Microtome (Micron, GMI, catalog number: 8243-30-0001)
Software
- Gen 5 TS 2.06 (BioTek Instruments Inc./BioTek, https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/)
- BD FACSDivaTM Software v8.0.3 (https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software)
- StepOne Software v2.3 (Thermo Fisher Scientific/Applied Biosystems, https://www.thermofisher.com/br/en/home/technical-resources/software-downloads/StepOne-and-StepOnePlus-Real-Time-PCR-System.html)
Procedure
- Surgical procedure
- Euthanize the rat using isoflurane according to the local regulations.
- Disinfect the rat by completely bathing with 1% iodized ethanol (Figure 1A) and wipe the abdomen and lower limbs with 2.5% chlorhexidine.
- Transfer the rat in a sterile surgical drape.
- Wear a sterile coat, mask, cap, and gloves.
- To prevent contamination, use sterile scissors and forceps, to make a small bilateral incision in the skin of the femorotibial joint region (Figure 1B).
- Use this incision as an access point to perform a bilateral divulsion toward the abdominal and inguinal region.
- Make a horizontal cut to join the two previously made incisions (Figure 1B).
- Using a #15 scalpel blade attached to cable #3, cut the patellar tendons, and the lateral and medial collateral ligaments bilaterally to expose the joint capsules.
- Perform joint capsule divulsion bilaterally.
- Remove the muscle tissue to expose the anterior part of the femur (Figure 1C).
- Cut the remaining ligaments and disarticulate the femoral hip joint.
- Remove the femur, and quickly clean off the majority of muscle and connective tissues attached to the bone.
- Transfer the femur to a 50-ml conical tube containing 15 ml of transport medium.
- Retract the skin from the abdominal and inguinal regions (Figure 1D).
- Carefully remove all adipose tissue without puncturing the abdominal wall and transfer it to a 50-ml conical tube containing 15 ml of transport medium.
- Take the conical tubes containing the fat tissue and femurs to the laminar flow hood.
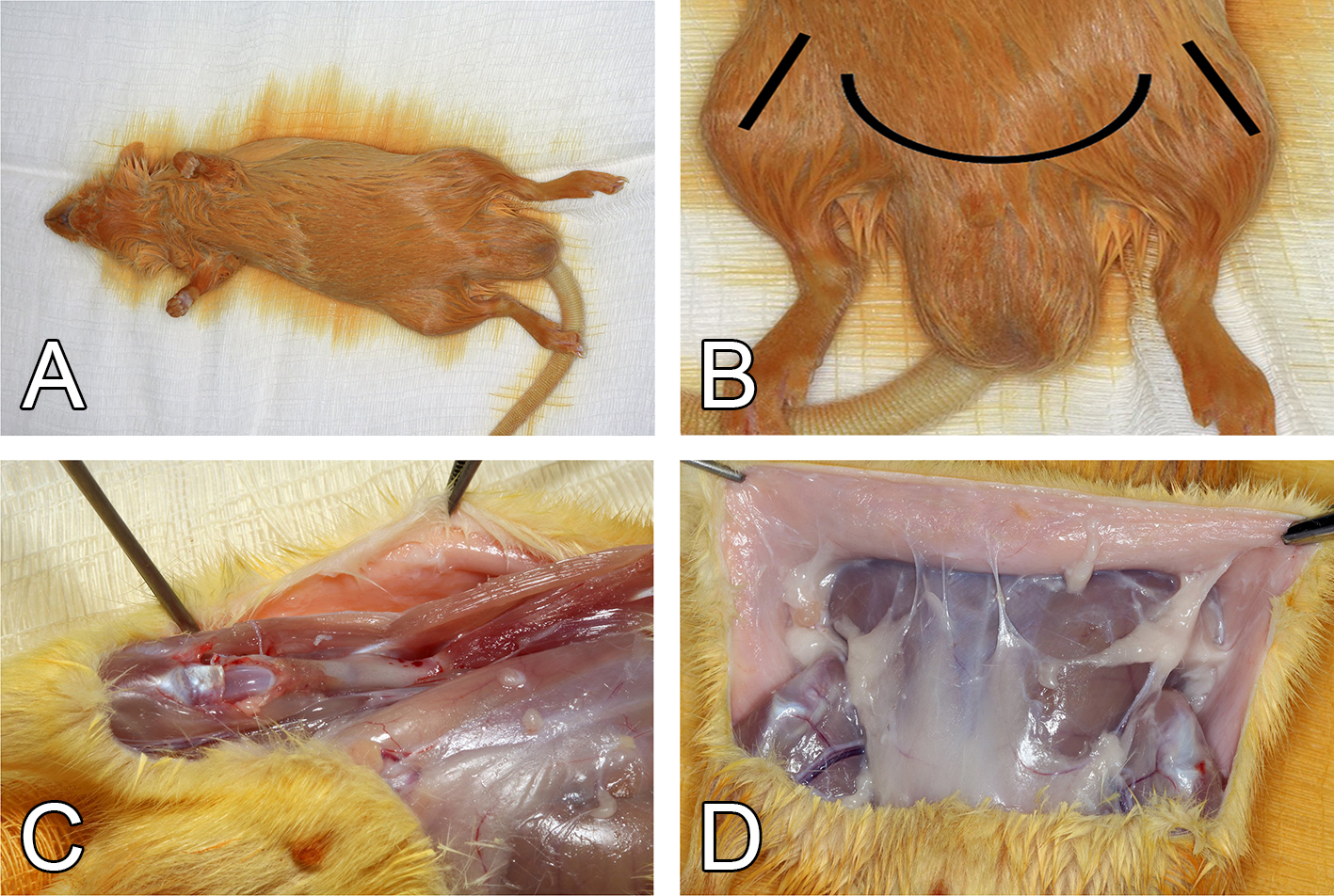
Figure 1. Surgical procedures for harvesting the femur and adipose tissue of a Wistar rat weighing 150-200 g. A. Disinfection of the animal with 1% iodized ethanol after euthanasia. B. Schematic representation of the incisions. C. Muscle removal and femur exposure. D. Skin retraction and exposure of the abdominal and inguinal adipose tissues.
- BM-MSC isolation and culture procedures
- Transfer the femurs from the conical tubes to a glass tissue culture dish filled with 70% ethanol.
- Within 1 min, remove the remaining connective tissue with sterile scissors, forceps and a #15 scalpel blade.
- Transfer the femurs to a new glass tissue culture dish filled with 2.5% chlorhexidine.
- Within 1 min, clean the remaining connective tissue with sterile scissors, forceps and a #15 scalpel blade (Figure 2A).
- Transfer the femurs to new conical tubes containing 15 ml of transport medium and incubate for 15 min at RT.
- Again, transfer the femurs to a new conical vial containing 15 ml transport medium and incubate for 15 min at RT.
- Lastly, transfer the femurs to a new conical vial containing 15 ml transport medium and incubate for 15 min at RT.
- Transfer the contents of this conical tube to a glass tissue culture dish.
- Fill a 20-ml syringe with growth medium and attach a 21G needle.
- Hold the femur with tweezers and cut the epiphyses using sterile scissors (Figure 2B).
- Insert the needle of the syringe filled with growth medium into the diaphysis and flush all bone marrow into a new 50-ml conical tube (Figure 2C).
- Centrifuge this tube for 5 min at 600 x g at RT.
- Discard the supernatant and resuspend the pellet in new growth medium (2 ml per femur).
- Transfer 2 ml of this suspension in a 75 cm2 cell culture flask filled with 10 ml of growth medium.
- Incubate this flask in an incubator at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air.
- After 24 h, gently rinse the flask three times with 1x PBS and replace with fresh growth medium.
- Change the culture medium every 2 days until the cells grow to 70% confluence.
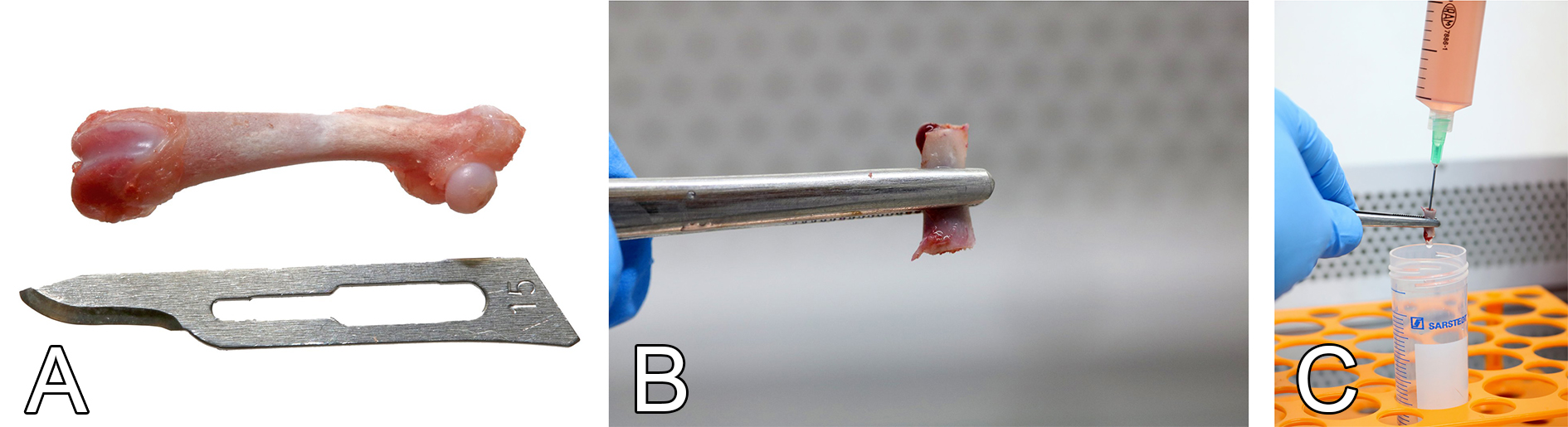
Figure 2. The harvesting of bone marrow from femur. A. Cleaned femur. B. Marrow cavity exposure after epiphyseal sectioning. C. Bone marrow flushing with growth medium using a needle and a syringe. - AT-MSC isolation and culture procedures
- Transfer the adipose tissue from the 50-ml conical tube to a glass tissue culture dish filled with 1x PBS to rinse the tissue.
- Transfer the adipose tissue to a new glass tissue culture dish (Figure 3A).
- Use sterile scissors to mince the adipose tissue into small pieces, around 1-2 mm3 (Figure 3B).
- Transfer the minced pieces to a 50-ml conical tube containing 20 ml of collagenase solution (Figure 3C).
- Place the tube in a water bath for 40 min at 37 °C, with shaking.
- Add 20 ml of growth medium to the 50-ml conical tube containing the adipose tissue and collagenase solution.
- Centrifuge the conical tube containing the adipose tissue, collagenase solution, and growth medium for 5 min at 600 x g.
- Discard the supernatant and resuspend the pellet in new growth medium (5 ml per adipose tissue removed from 1 animal).
- Transfer 5 ml of this suspension into a 75 cm2 cell culture flask filled with 10 ml of growth medium.
- Incubate this flask in an incubator at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air.
- After 24 h, gently wash the flask three times with 1x PBS and replace with fresh growth medium.
- Replace the culture medium every 2 days until cells are 70% confluent.
- After 7 days of culture in growth medium, approximately 5 x 106 MSCs were generated from the adipose tissue of each animal.
- Typically, within 7 days, MSCs reach 70% of confluence and are ready to be used in subsequent experiments (Figure 4).
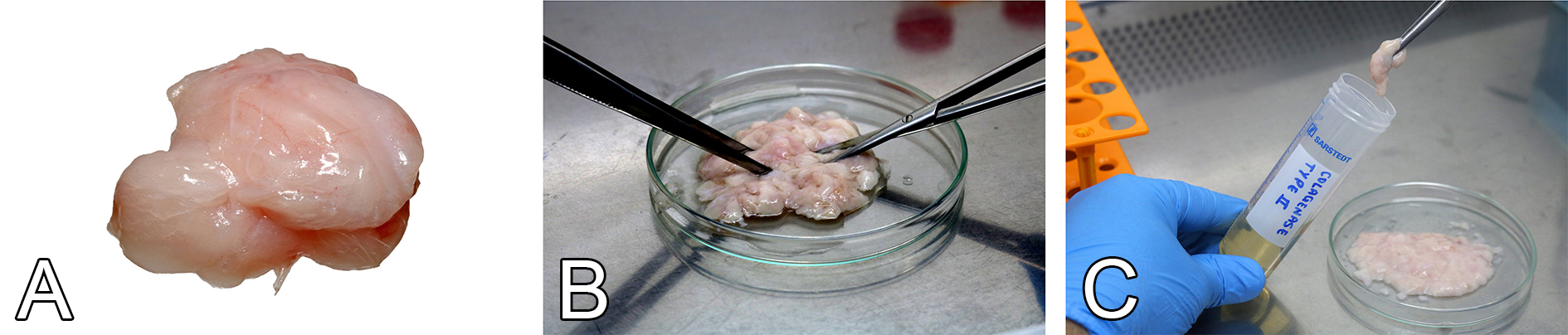
Figure 3. The enzymatic digestion of abdominal and inguinal adipose tissue. A. Harvested adipose tissue. B. Mincing of adipose tissue into small pieces using sterile scissors and tweezers. C. Transfer adipose tissue pieces to collagenase type II solution for enzymatic digestion and cell isolation.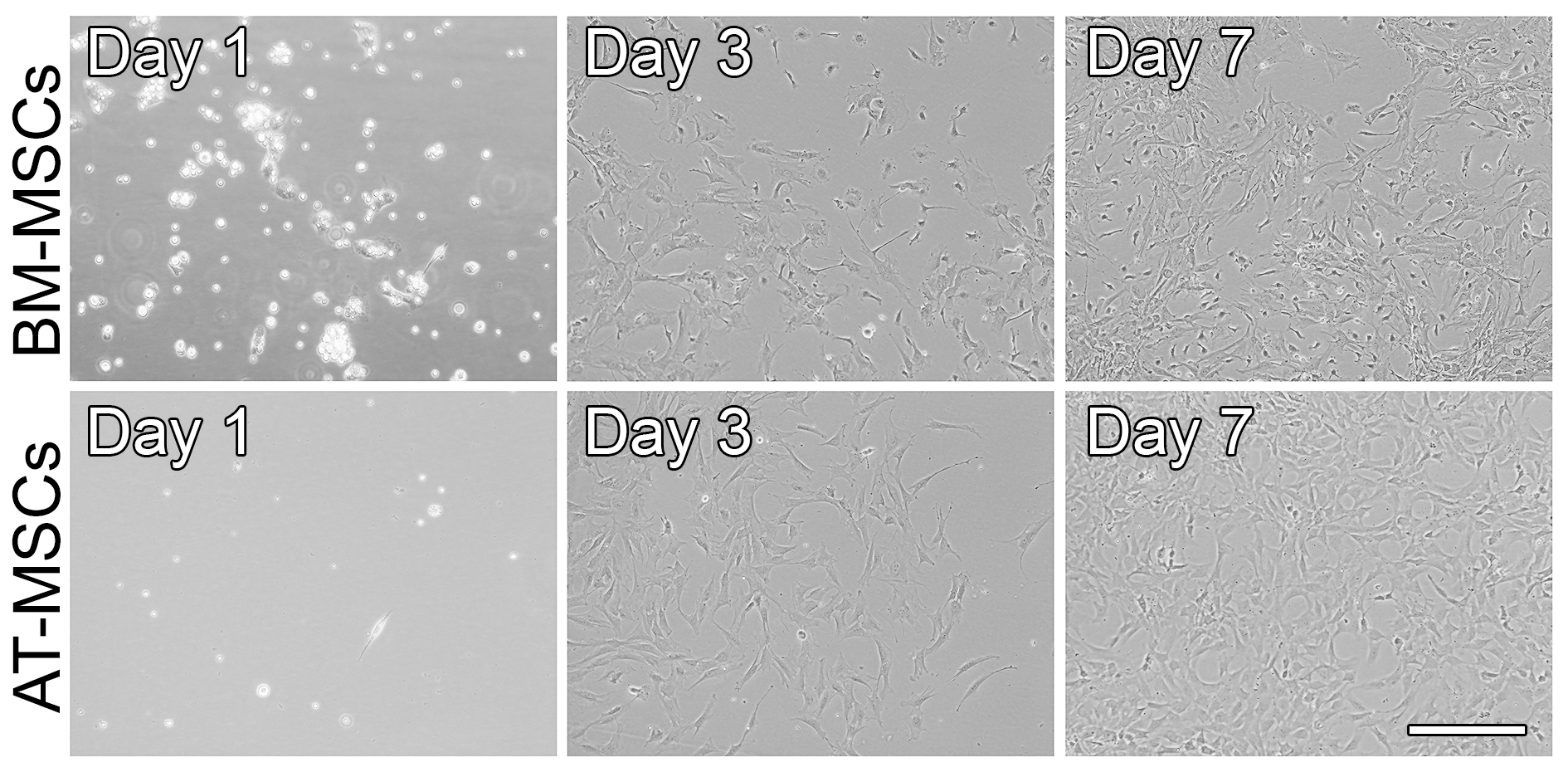
Figure 4. Phase-contrast micrographs showing the morphology of BM-MSCs and AT-MSCs that were cultured in growth medium and on polystyrene dishes for up to 7 days. After 24 h, both BM-MSCs and AT-MSCs have attached to the polystyrene dish, and their morphology is round/oval. As the cells were cultured, they proliferated and became elongated, polygonal, and spindle-shaped. Scale bar = 100 µm. - Characterization of BM-MSCs and AT-MSCs
- Wash the flask three times with 1x PBS.
- Add 5 ml of trypsin solution into the flask and incubate for 5 min at 37 °C.
- Add 2.5 ml of fresh growth medium into the flask, transfer the cell suspension into a 50-ml conical tube, and centrifuge for 5 min at 600 x g.
- Discard the supernatant.
- Wash the cell pellet once with 1x PBS.
- Centrifuge the cell suspension for 5 min at 600 x g.
- Discard the supernatant.
- Add 5 ml of 1x PBS to the cell pellet and mix the cell suspension.
- Count the cells in a hemocytometer (Neubauer Chamber).
- Adjust the concentration of the cell suspension to obtain a density of 2 x 105 cells/ml with 1x PBS.
- Add 1 ml of cell suspension to each flow cytometer tube (one tube for each specific antibody, one tube for isotype control, and one tube with cells that will not be labeled with antibody).
- Centrifuge the flow cytometer tubes for 5 min at 600 x g.
- Discard the supernatant.
- Add 100 µl of 1x PBS to the cell pellet and mix by flicking/tapping the tube.
- Incubate each tube for 30 min at RT in the dark with 2 µl of the following monoclonal anti-rat antibodies: anti-CD29, -CD31, -CD34, -CD45, and -CD106, directly conjugated with a fluorophore (antibody final dilution: 1:50). For monoclonal anti-rat antibody -CD90 directly conjugated with a fluorophore: dilute the antibody 1:5 in 1x PBS and then add 2 µl to the cell suspension (antibody final dilution: 1:250).
- Add 2 µl of isotype control to the corresponding tube.
- Wash the cells with 2 ml of 1x PBS.
- Centrifuge for 5 min at 600 x g.
- Discard the supernatant.
- Add 0.5 ml of formaldehyde solution (4%) diluted to 1% in 1x PBS.
- Analyze the cells by flow cytometry (Figures 5 and 6).
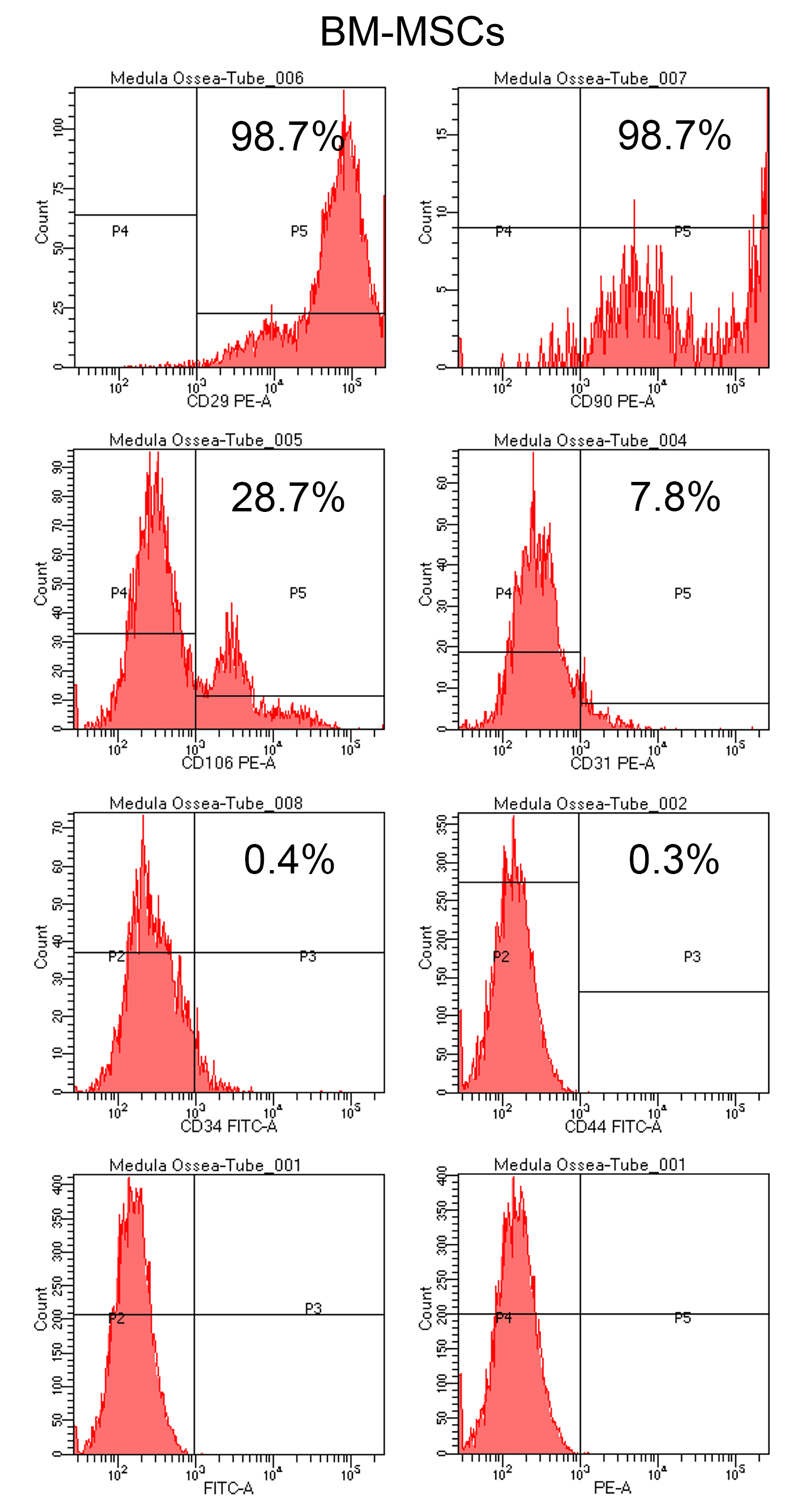
Figure 5. Flow cytometry analysis of BM-MSCs cultured in growth medium on a polystyrene culture dish for 7 days. Histograms show the expression of the surface markers CD29, CD90, CD106, CD31, CD34, and CD44 after incubation with the respective antibodies. Cells were also incubated with the isotypes FITC-A and PE-A, which were used as negative controls. A high percentage of BM-MSCs expressed CD29, CD90, and CD106 (98.7%, 98.7%, and 28.7%, respectively) and a low percentage expressed CD31, CD34, and CD44 (7.8%, 0.4%, and 0.3%, respectively).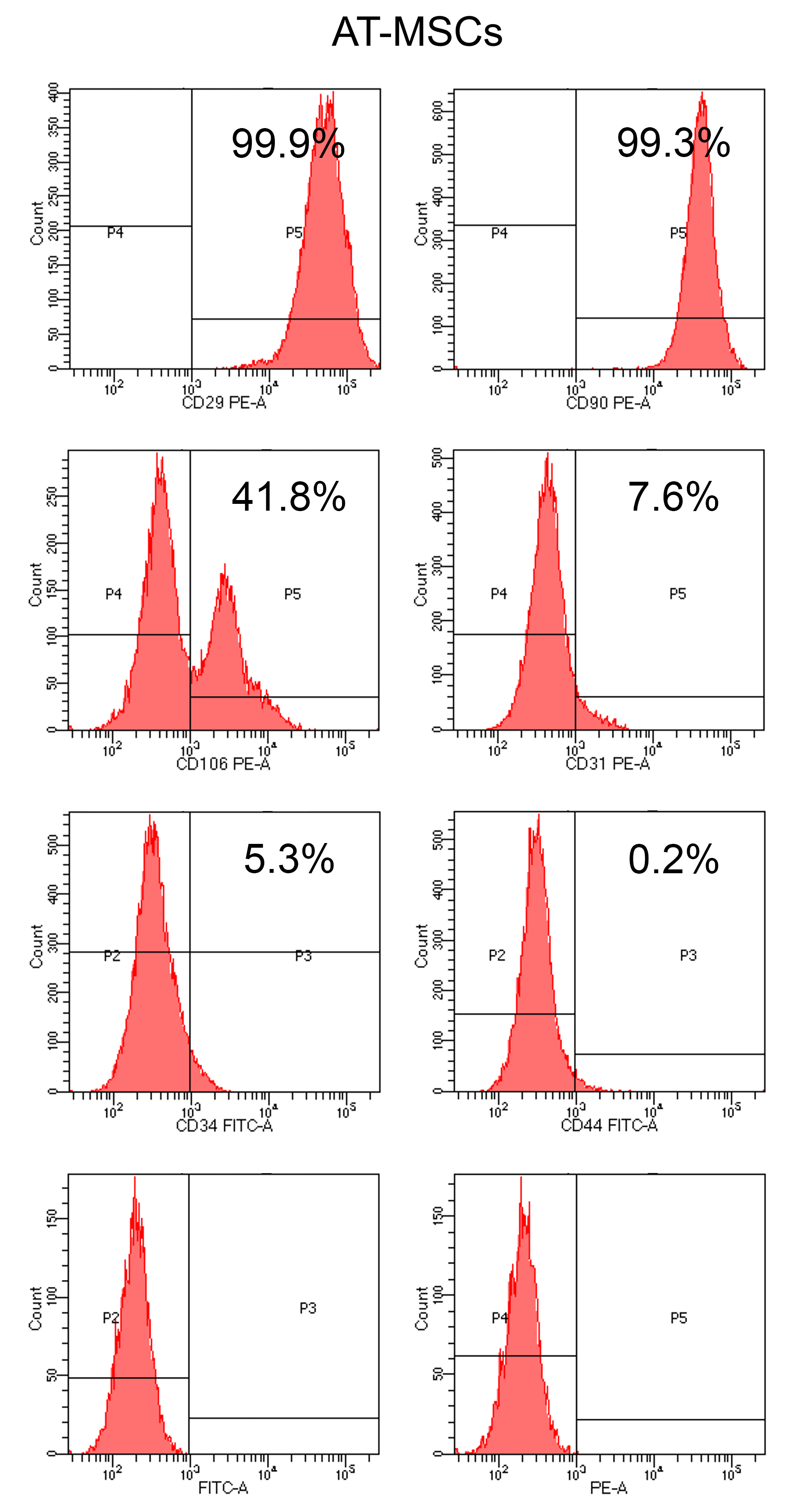
Figure 6. Flow cytometry analysis of AT-MSCs cultured in growth medium on a polystyrene dish for 7 days. Histograms show the expression of the surface markers CD29, CD90, CD106, CD31, CD34, and CD44 after incubation with the respective antibodies. Cells were also incubated with the isotypes FITC-A and PE-A, which were used as negative controls. A high percentage of AT-MSC expressed CD29, CD90, and CD106 (99.9%, 99.3%, and 41.8%, respectively) and a low percentage expressed CD31, CD34, and CD44 (7.6%, 5.3%, and 0.2%, respectively).
- Osteoblast differentiation
- When BM-MSCs or AT-MSCs reach 70% confluence, remove the growth medium.
- Wash the flask three times with 1x PBS.
- Add 5 ml of trypsin solution into the flask and incubate for 5 min at 37 °C.
- Add 2.5 ml of fresh growth medium into the flask, transfer the cell suspension to a 50-ml conical tube, and centrifuge for 5 min at 600 x g.
- Discard the supernatant and resuspend the cell pellet in new growth medium.
- Count the cells and plate them at a cell density of 2 x 104 cells/well in 24-well culture plates in 1 ml of osteogenic medium or 1 x 105 cells/well in 6-well culture plates in 2 ml of osteogenic medium.
- Incubate the plates in an incubator at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air during the time-course of the experiment.
- Replace the culture medium every 2 days.
- The extracellular matrix mineralization can be observed after 21 days in culture.
- Alizarin red staining
To confirm osteoblast differentiation, one of the methods we used is the detection of mineralized extracellular matrix by alizarin red staining.- Remove the culture medium from each well and gently wash the cells 3 times with 1x PBS.
- Add 10% formalin and incubate at 4 °C for 24 h (24-well plates–500 µl; 12-well plates–1 ml; 6-well plates–2.4 ml).
- Remove the 10% formalin and dehydrate the cells using increasing concentrations of ethanol (30%, 50%, 70%, and 96%) for 1 h each (24-well plates–500 µl; 12-well plates–1 ml; 6-well plates–2.4 ml).
- Remove the 96% ethanol and incubate at RT until the wells are dry.
- Cover the well with alizarin red staining and incubate at RT for 10 min.
- Wash once with deionized water and incubate at RT until the wells are dry.
- Take macroscopic (Figure 7) and microscopic photos of the wells.
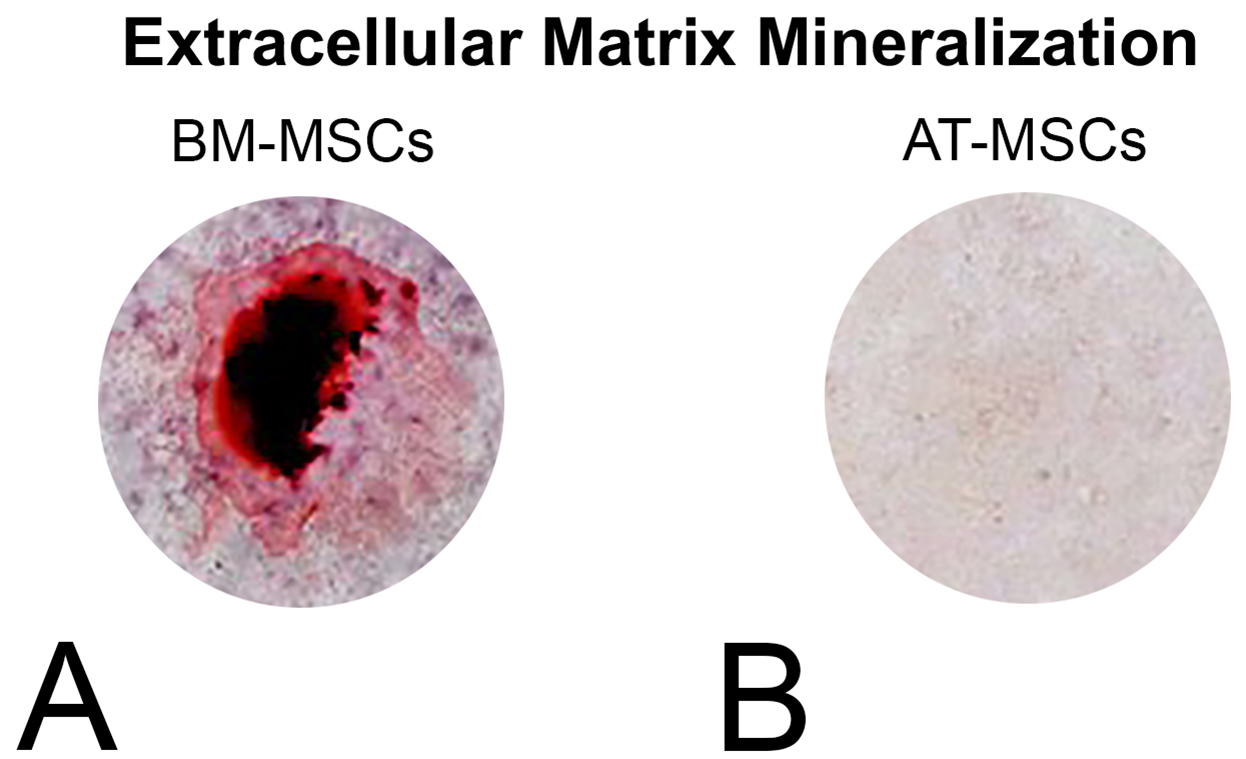
Figure 7. Mineralized extracellular matrix detected by alizarin red staining in BM-MSC and AT-MSC cultures after 21 days of culture in osteogenic medium on polystyrene dishes
- Chondroblast differentiation
- When BM-MSC and AT-MSC cultures reach 70% confluence, remove the growth medium.
- Wash the flask three times with 1x PBS.
- Add 5 ml of trypsin solution into the flask and incubate for 5 min at 37 °C.
- Add 2.5 ml of fresh growth medium into the flask.
- Transfer this cell suspension to a 50-ml conical tube and centrifuge for 5 min at 600 x g.
- Discard the supernatant.
- Resuspend the cells in chondroblast differentiation medium at a density of 1.25 x 106 cells/ml.
- Using a pipette, dispense 200 µl aliquots of the cell suspension (2.5 x 105 cells) into each well of polypropylene 96-well plates.
- Centrifuge the plates at 500 x g for 5 min.
- Add 200 μl of 1x PBS into the empty wells to minimize the evaporation of the culture medium.
- Incubate the plates in an incubator at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air during the time-course of the experiment.
- After 24 h of incubation, cell aggregates were visible, and the chondrogenic phenotype was observed after 30 days in culture.
- Replace the culture medium every 2 days by carefully aspirating the expired medium using a sterile 200 μl pipette and adding 200 μl of fresh chondrogenic medium to each well.
- Trichrome staining
To confirm chondroblast differentiation, one of the methods we used is the detection of collagen fibers with trichrome staining.- Using a micropipette, remove the chondroblast differentiation medium.
- Add 200 μl of 1x PBS into each well.
- Using a micropipette, remove the PBS.
- Add 200 μl of 4% paraformaldehyde for 5 min at RT.
- Remove the paraformaldehyde.
- Wash each well with 1x PBS, 2 times for 3 min each.
- Stain the cell aggregates with eosin for 5 min at RT.
- Remove the eosin and wash with 1x PBS, 2 times for 3 min each.
- Using a 1,000 µl micropipette, harvest the aggregates and transfer them to 1.5-ml microtubes.
- Dehydrate the cells aggregates in 300 µl of a graded ethanol series (70%, 80%, 90%, 95%, and 100%, 5 min each) by placing them into 1.5-ml microtubes.
- Remove the 100% ethanol and perform three clarification steps in 300 µl xylene for 3 min each.
- Paraffin-embed the aggregates into a mold in a hot surface for 5 min then transfer each mold to a cold surface.
- Cut adjacent 5 μm sections using a microtome.
- Deparaffinize the sections overnight in an incubator at 60 °C.
- Deparaffinize the sections in three steps of xylene for 5 min each, and rehydrate the sections by incubation in an ethanol series (100%, 95%, 90%, 80%, and 70%, 3 min each).
- Wash the sections with deionized water for 5 min.
- Stain with acid fuchsin (HT15-1) for 5 min at RT.
- Wash with water for 5 min.
- Stain with a working solution of phosphomolybdic acid (HT15-3) and phosphotungstic acid (HT15-2) for 5 min at RT.
- Stain the sections with an aniline blue solution for 5 min at RT.
- Remove the excess aniline blue, and add a 1% acetic acid solution for 3 min.
- Rinse with sections with tap water for 3 min.
- Dehydrate sections in a graded ethanol series (70%, 80%, 90%, 95%, and 100%, 1 min each), followed by three clarification steps in xylene for 1 min each.
- Mount the slides using Erv-Mount®.
- Take microscopic photos of the histological sections (Figure 8).
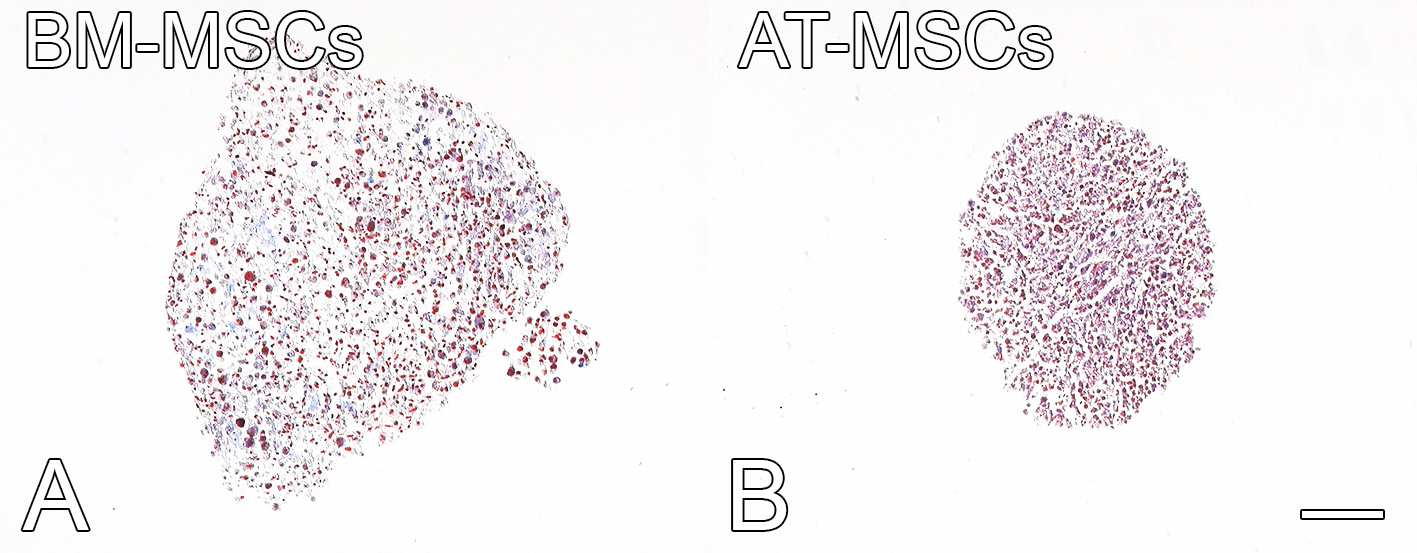
Figure 8. Collagen fibers (blue) and cytoplasm (red) detected by trichrome staining in BM-MSCs and AT-MSCs cultured in chondrogenic medium on ultra-low cluster 96-well plates for 30 days. Scale bar = 100 µm. - Adipocyte differentiation
- When BM-MSC and AT-MSC cultures reach 70% confluence, remove the growth medium.
- Wash the flask three times with 1x PBS.
- Add 5 ml of trypsin solution into the flask and incubate for 5 min at 37 °C.
- Add 2.5 ml of fresh growth medium into the flask.
- Transfer this cell suspension to a 50-ml conical tube and centrifuge for 5 min at 600 x g.
- Discard the supernatant and resuspend the cell pellet in fresh growth medium.
- Count the cells and plate them at a cell density of 2 x 104 cells/well in 24-well culture in 1 ml of adipogenic medium plates or 1 x 105 cells/well in 6-well culture plates in 2 ml of adipogenic medium.
- Incubate the plates in an incubator at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air during the time-course of the experiment.
- Replace the culture medium every 2 days. The intracytoplasmic lipid droplets were observed after 10 days in culture.
- Oil red O staining
To confirm adipocyte differentiation, one of the methods we used is the detection of intracytoplasmic lipid droplets with oil red O staining.- Remove the culture medium from each well.
- Add 10% formalin and incubate for 5 min at RT (24-well plates–500 µl, 12-well plates–1 ml, 6-well plates–2.4 ml).
- Discard the 10% formalin and add the same volume of fresh 10% formalin. Incubate for at least 1 h.
Note: Cells can be kept in formalin for a couple of days before staining. Wrap parafilm around the plate to prevent the cells from drying out and cover the plate with aluminum foil. - Remove the 10% formalin using a small transfer pipette.
- Wash the wells with 60% isopropanol (24-well plates–500 µl, 12-well plates–1 ml, 6-well plates–2.4 ml).
- Let the wells dry completely.
- Add the oil red O staining working solution for 10 min (do not touch walls of the wells).
- Remove the oil red O staining, and immediately add deionized water (repeat this step 4 times).
- Remove all deionized water and incubate at RT to dry.
- Image the wells using a phase-contrast microscope (Figure 9).
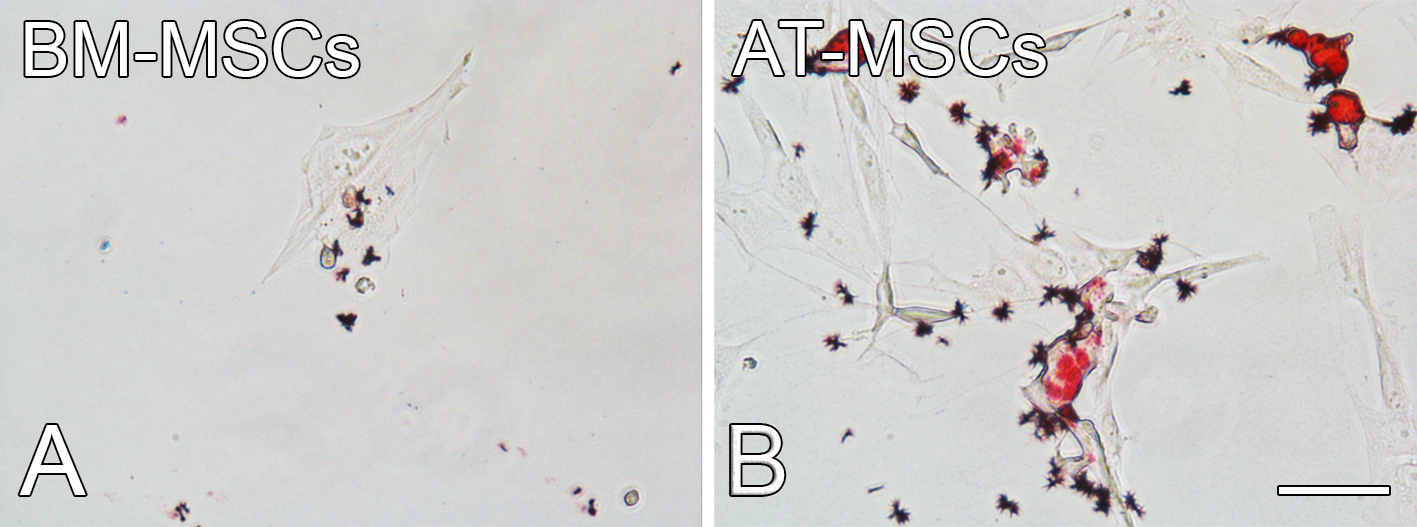
Figure 9. Intracytoplasmic lipid droplets detected by oil red O staining in BM-MSCs and AT-MSCs cultured in adipogenic medium on a polystyrene dish for 10 days. Scale bar = 100 µm.
Recipes
- Transport medium
Note: Prepared fresh just prior to use and kept at 37 °C.
57 ml of alpha minimum essential medium (α-MEM)
3 ml of gentamycin (50 µg/ml)
720 µl of amphotericin B (0.3 µg/ml) - Collagenase solution
Note: Prepared fresh just prior to use and placed at RT.
15 mg of type II collagenase (0.075%)
20 ml of 1x PBS
Filter this solution in the laminar flow hood into a 50-ml conical tube using a 20-ml syringe and 0.2 µm filter - Trypsin solution
Note: Prepared fresh just prior to use and placed at RT.
19 ml of trypsin (0.25%)
500 µl of type II collagenase (1.3 mg/ml)
1 ml of EDTA (1 mM) - Ascorbic acid and β-Glycerophosphate solution
Note: Previously prepared, kept at 4 °C for up to 7 days.
5 mg of ascorbic acid (5 µg/ml)
2.16 g of β-glycerophosphate (7 mM)
10 ml of deionized water
Filter this solution in the laminar flow hood into a 50-ml conical tube using a 20-ml syringe and 0.2 µm filter - Growth medium (10% MEM)
Note: Previously prepared, kept at 4 °C for up to 30 days.
360 ml of α-MEM
40 ml of fetal calf serum
2 ml of gentamycin (50 µg/ml)
500 µl of amphotericin B (0.3 µg/ml) - Osteogenic differentiation medium
Note: Previously prepared, kept at 4 °C for up to 30 days.
400 ml of growth medium
4 ml of dexamethasone (10-7 M)
1% ascorbic acid and β-glycerophosphate solution
Note: The ascorbic acid and β-glycerophosphate solution are added immediately before using the medium. - Chondrogenic differentiation medium
Note: Previously prepared, kept at 4 °C for up to 30 days.
100 ml of D-MEM
100 µl of sodium pyruvate (100 mM)
100 µl of dexamethasone (1 mM)
250 µl of ascorbic acid (20 mM)
1 ml of human albumin (0.02%)
20 µl/ml of transforming growth factor β3 (TGF-β3, 1 µg/ml).
Note: The TGF-β3 is added immediately before using the medium. - Adipocyte differentiation medium
Note: Previously prepared, kept at 4 °C for up to 30 days.
180 ml of D-MEM
20 ml of fetal calf serum (10%)
2 ml of gentamycin (50 µg/ml)
250 µl of amphotericin B (0.3 µg/ml)
2 ml of dexamethasone (10-6 M)
2 ml of 3-isobutyl-1-methylxanthine (0.5 mM)
260 µl of indomethacin (0.1 M)
150 µl of insulin (10 mg/ml) - Dexamethasone stock solution (200 µM)
Note: Previously prepared, kept at -20 °C.
Dexamethasone is dissolved at 200 µM in absolute ethanol and deionized water - Ascorbic acid stock solution (20 mM)
Note: Previously prepared, kept at 4 °C for up to 7 days.
A stock solution of 20 mM ascorbic acid is prepared in 1x PBS - TGF-β3
Note: Prepared fresh just prior to use and placed at RT.
A 1 μg/ml stock solution of TGF-β3 is prepared in 1x PBS and 0.5% human albumin. - Oil red O staining
Oil red O stock solution
Note: Previously prepared, kept at RT up to 6 months.
Oil red O (700 mg) is added to 200 ml of isopropanol, stirred overnight, and then passed through a 0.2 µm filter.
Oil red O work solution
Mix 6 parts of oil red O stock solution with 4 parts deionized water and incubate at RT for 20 min.
Acknowledgments
This study was funded by the State of Sao Paulo Research Foundation (FAPESP, Brazil, #2017/12622-7), National Council for Scientific and Technological Development (CNPq, Brazil, # 305523/2013-9 and 404318/2016-9), and Coordination of Improvement of Higher Education Personnel (CAPES, Brazil). The English language review was carried out by ENAGO (www.enago.com) funded by FAPESP (#2018/17356-6).This protocol was adapted from these works (Maniatopoulos et al., 1988; Huang et al., 2002; Penick et al.,2005).
Competing interests
The authors declare no conflict of interest.
Ethics
All procedures performed were conducted in accordance with the ethical standards of the international, national, and/or institutional animal care guidelines. The Committee of Ethics in Animal Research of the School of Dentistry of Ribeirão Preto, University of São Paulo (#2018.1.30.58.8) reviewed and approved all animal procedures we have done here.
References
- Abuna, R. P., De Oliveira, F. S., Santos Tde, S., Guerra, T. R., Rosa, A. L. and Beloti, M. M. (2016). Participation of TNF-α in inhibitory effects of adipocytes on osteoblast differentiation. J Cell Physiol 231(1): 204-214.
- Almeida, A. L. G., Freitas, G. P., Lopes, H. B., Gimenes, R., Siessere, S., Sousa, L. G., Beloti, M. M. and Rosa, A. L. (2019). Effect of stem cells combined with a polymer/ceramic membrane on osteoporotic bone repair. Braz Oral Res 33: e079.
- Bianco, P., Robey, P. G. and Simmons, P. J. (2008). Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell 2(4): 313-319.
- Caplan, A. I. (1991). Mesenchymal stem cells. J Orthop Res 9(5): 641-650.
- Fideles, S. O. M., Ortiz, A. C., Assis, A. F., Duarte, M. J., Oliveira, F. S., Passos, G. A., Beloti, M. M. and Rosa, A. L. (2019). Effect of cell source and osteoblast differentiation on gene expression profiles of mesenchymal stem cells derived from bone marrow or adipose tissue. J Cell Biochem 120(7): 11842-11852.
- Freitas, G. P., Lopes, H. B., Souza, A. T. P., Oliveira, P., Almeida, A. L. G., Souza, L. E. B., Coelho, P. G., Beloti, M. M. and Rosa, A. L. (2019). Cell therapy: effect of locally injected mesenchymal stromal cells derived from bone marrow or adipose tissue on bone regeneration of rat calvarial defects. Sci Rep 9(1): 13476.
- Friedenstein, A. J., Chailakhjan, R. K. and Lalykina, K. S. (1970). The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet 3(4): 393-403.
- Harting, M., Jimenez, F., Pati, S., Baumgartner, J. and Cox, C., Jr. (2008). Immunophenotype characterization of rat mesenchymal stromal cells. Cytotherapy 10(3): 243-253.
- Hu, Q., Liu, M., Chen, G., Xu, Z. and Lv, Y. (2018). Demineralized bone scaffolds with tunable matrix stiffness for efficient bone integration. ACS Appl Mater Interfaces 10(33): 27669-27680.
- Huang, J. I., Beanes, S. R., Zhu, M., Lorenz, H. P., Hedrick, M. H., Benhaim, P. (2002). Rat extramedullary adipose tissue as a source of osteochondrogenic progenitor cells. Plast Reconstr Surg 109(3):1033-1041.
- Lopes, H. B., Freitas, G. P., Fantacini, D. M. C., Picanço-Castro, V., Covas, D. T., Rosa, A. L. and Beloti, M. M. (2019). Titanium with nanotopography induces osteoblast differentiation through regulation of integrin αV. J Cell Biochem 120(10): 16723-16732.
- Maniatopoulos, C., Sodek, J., Melcher, A. H. (1988). Bone formation in vitro by stromal cells obtained from bone marrow of young adult rats. Cell Tissue Res 254(2): 317-330.
- Nancarrow-Lei, R., Mafi, P., Mafi, R. and Khan, W. (2017). A systemic review of adult mesenchymal stem cell sources and their multilineage differentiation potential relevant to musculoskeletal tissue repair and regeneration. Curr Stem Cell Res Ther 12(8): 601-610.
- Oliveira, F. S., Bellesini, L. S., Defino, H. L., da Silva Herrero, C. F., Beloti, M. M. and Rosa, A. L. (2012). Hedgehog signaling and osteoblast gene expression are regulated by purmorphamine in human mesenchymal stem cells. J Cell Biochem 113(1): 204-208.
- Penick, K. J., Solchaga, L. A. and Welter, J. F. (2005). High-throughput aggregate culture system to assess the chondrogenic potential of mesenchymal stem cells. Biotechniques 39(5): 687-691.
- Pierini, M., Dozza, B., Lucarelli, E., Tazzari, P. L., Ricci, F., Remondini, D., di Bella, C., Giannini, S. and Donati, D. (2012). Efficient isolation and enrichment of mesenchymal stem cells from bone marrow. Cytotherapy 14(6): 686-693.
- Schrepfer, S., Deuse, T., Lange, C., Katzenberg, R., Reichenspurner, H., Robbins, R. C. and Pelletier, M. P. (2007). Simplified protocol to isolate, purify, and culture expand mesenchymal stem cells. Stem Cells Dev 16(1): 105-107.
- Wadajkar, A. S., Santimano, S., Tang, L. and Nguyen, K. T. (2014). Magnetic-based multi-layer microparticles for endothelial progenitor cell isolation, enrichment, and detachment. Biomaterials 35(2): 654-663.
- Zhang, Y. D., Zhao, S. C., Zhu, Z. S., Wang, Y. F., Liu, J. X., Zhang, Z. C. and Xue, F. (2017). Cx43- and smad-mediated TGF-beta/BMP signaling pathway promotes cartilage differentiation of bone marrow mesenchymal stem cells and inhibits osteoblast differentiation. Cell Physiol Biochem 42(4): 1277-1293.
- Zuk, P. A., Zhu, M., Mizuno, H., Huang, J., Futrell, J. W., Katz, A. J., Benhaim, P., Lorenz, H. P. and Hedrick, M. H. (2001). Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 7(2): 211-228.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Freitas, G. P., Souza, A. T. P., Lopes, H. B., Trevisan, R. L. B., Oliveira, F. S., Fernandes, R. R., Ferreira, F. U., Ros, F. A., Beloti, M. M. and Rosa, A. L. (2020). Mesenchymal Stromal Cells Derived from Bone Marrow and Adipose Tissue: Isolation, Culture, Characterization and Differentiation. Bio-protocol 10(4): e3534. DOI: 10.21769/BioProtoc.3534.
Category
Stem Cell > Adult stem cell > Adipose Stem Cell
Stem Cell > Adult stem cell > Hematopoietic stem cell
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link