- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Transcervical Mouse Infections with Chlamydia trachomatis and Determination of Bacterial Burden
Published: Vol 10, Iss 3, Feb 5, 2020 DOI: 10.21769/BioProtoc.3506 Views: 5922
Reviewed by: Alka MehraFrank FollmannAchille Broggi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

PBMC-Humanized Mouse Model for Multiple Sclerosis: Studying Immune Changes and CNS Involvement
Anastasia Dagkonaki [...] Lesley Probert
May 20, 2025 4018 Views

In Vitro Co-culture of Bacterial and Mammalian Cells to Investigate Effects of Potential Probiotics on Intestinal Barrier Function
Ajitpal Purba [...] Dulantha Ulluwishewa
Jun 20, 2025 2589 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1689 Views
Abstract
Chlamydia trachomatis is an obligate human pathogen. It infects the genital tract of humans ascending into the fallopian tube, exacerbated by chronic pelvic pain, pelvic inflammatory disease, and fallopian tube scaring resulting in infertility and other malignancies. The major hurdle in controlling chlamydial spread is that the infection remains asymptomatic, thus leading to chronic, recurrent and persistent infections, with no vaccines developed so far. Being a human pathogen, we do not have an in vivo model of C. trachomatis infection. C. trachomatis do not cause ascending infections and fallopian tube pathology in the mouse urogenital tract when infected vaginally. To overcome this hurdle trans cervical method of infection must be adapted. In this protocol the method of establishing trans-cervical chlamydial infection with the procedure to determine the bacterial load is detailed. This method will facilitate to deliver the bacteria past the cervix establishing an ascending infection into the uterine horns reciprocating human fallopian tube infections.
Keywords: Chlamydia trachomatisBackground
Sexually transmitted infections (STI) remain major societal and economic burden despite public health initiatives, vaccinations and development of antibiotics (O’Connell and Ferone, 2016). There is estimated to be 357 million new infections annually and STIs have been identified by the WHO as a global priority for elimination (WHO Global Health Sector Strategy on Sexually Transmitted Infections 2016-2021). Among the various bacterial STI, Chlamydia trachomatis is the most common STI (Starnbach, 2018) and stands first with a 131 million new infections annually worldwide (Poston et al., 2017). High reinfection in the genital tract is exacerbated by chronic pelvic pain, pelvic inflammatory disease, and fallopian tube scaring (O’Connell and Ferone, 2016) resulting in infertility and other malignancies. The major hurdle in controlling chlamydial spread is that the infection remains asymptomatic, thus leading to chronic, recurrent and persistent infections, with no vaccines developed so far.
Chlamydia undergoes a unique, biphasic developmental cycle that generally modulates between two morphological forms. Extracellular, infectious elementary bodies (EBs) attach to host cells within 15 min of infection, after which they are internalized into a membrane bound vesicle called an inclusion. EBs then differentiate into metabolically active, non-infectious, reticulate bodies (RBs) in 12 h that undergo binary fission, followed by secondary differentiation into EBs after 36 h post infection (hpi). Within/after 48 hpi most of the bacteria is in the EB form and are released upon host cell lysis or extrusion (Abdelrahman and Belland, 2005).
Chlamydia trachomatis being an obligate human pathogen lacks an in vivo model. To evaluate specific vaccine antigens that can be used in humans, and to study the host pathogen interaction to understand the pathogenicity of Chlamydia infection, a suitable in vivo model need to be established (De Clercq et al., 2013). Unlike in Chlamydia muridarium, a natural pathogen in mouse, intravaginal inoculation of the bacteria only leads to a mild genital infection and does not cause ascending infection or tubal pathology. The innate immune response in the lower genital tract of the mouse can rapidly eliminate human chlamydial organism (Sturdevant and Caldwell, 2014). Thus a higher number of infectious particles are required to establish infection, which yields a 2 log units lower bacterial load in the uterine horns (Darville et al., 1997; Ramsey et al., 2000). To address this limitation an intra-bursal mode of infection was established (Carmichael et al., 2013), via a survival surgery and directly inoculating the bacteria into the bursal region. This method bypasses both the cervical barrier and endometrial lumen, thus do not resemble the natural path of vaginal infection (see Figure 1).
The primary site of Chlamydia trachomatis infection is the cervical epithelium (Buckner et al., 2016). Subsequently, the infection ascends to the upper genital tract tissues, the uterine horns and oviducts, which frequently lead to hydrosalpinx, fibrosis, and infertility, which are also common post-infection sequelae in women (Morrison and Caldwell, 2002; Shah et al., 2005). Mice have a bicornuate uterus with two lateral horns and are lined with ligaments carrying blood and lymphatic vessels. The cervix has cranial, fundal and caudal segments. The caudal segment or cervix consists of a single cavity that protrudes into the vaginal opening. To establish an ascending infection, which closely resembles infection in human uterus and fallopian tube, we need to bypass the cervix and inoculate the pathogen into the uterine fundus. This leads to an easy establishment of ascending infection in mouse. This method has been made used in various study (Gondek et al., 2012; Pal et al., 2018; Rajeeve et al., 2018).
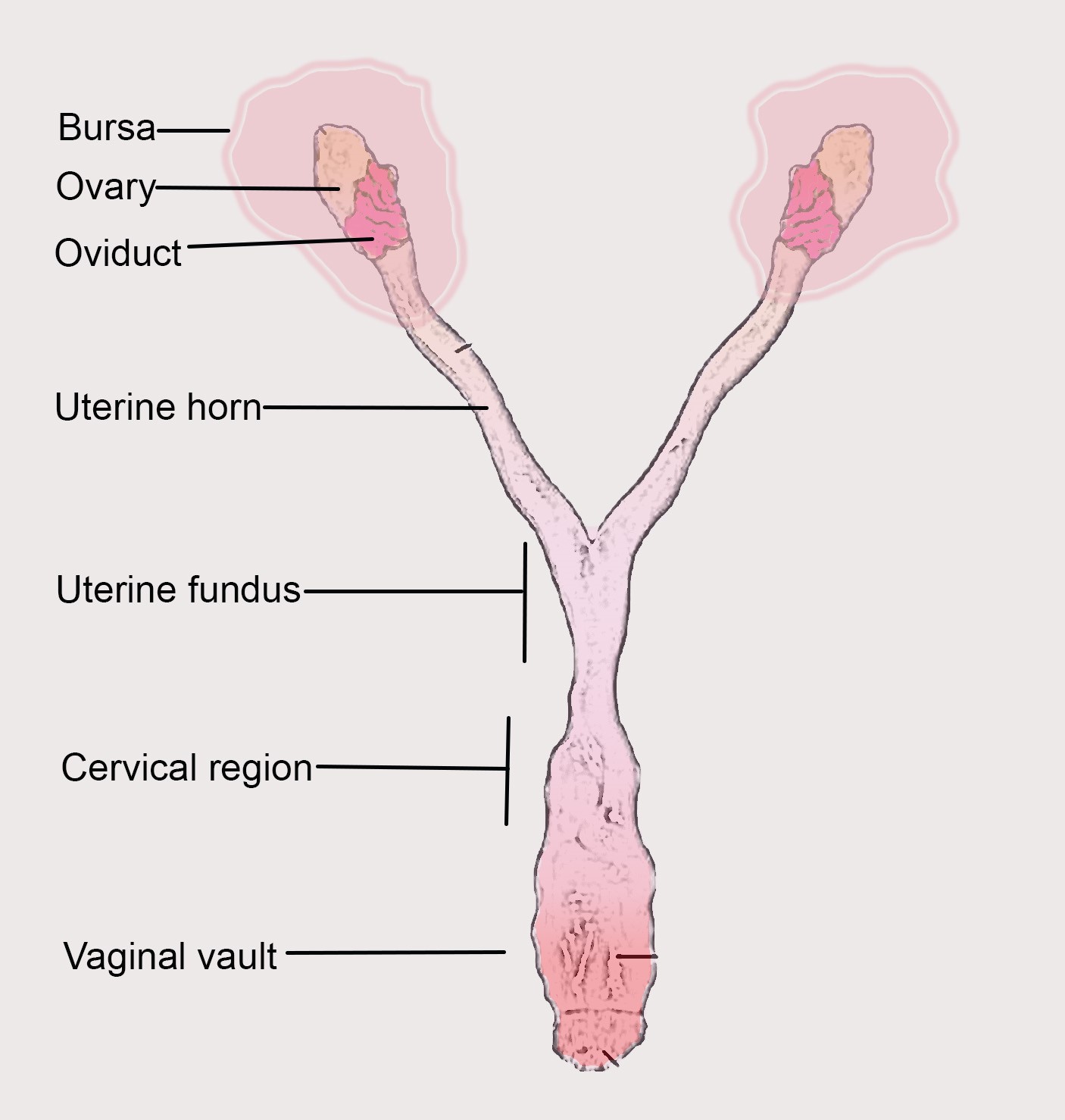
Figure 1. Mouse reproductive tract with parts labelled
Materials and Reagents
Transcervical infection and DNA extraction
- Eppendorf tubes (Stastedt GmbH, catalog number: 72.688.004)
- Ultracentrifugation tubes (Beckman Coulter, catalog number: NC9146666)
- 15 ml Falcons (Corning, catalog number: 430791)
- 50 ml Falcon
- 12-well and 24-well plate
- Rubber scraper
- 75 cm2 or 150 cm2 flasks (Corning, catalog numbers: CLS3290 and CLS3291)
- Nitrile glove (Microflex Neotouch, catalog number: 93-833)
- Sterile 200 μl pipette tips (Stastedt GmbH, catalog number: 25-201)
- 1 ml syringe (BD Leur-LokTM, catalog number: 309628)
- NSET: Non-surgical embryo transfer device (ParaTechs Corp, Product number: 60010)
- Post-pubertal and sexually mature (> 42 days old) female mice (any strain)
- Medroxy-progesterone acetate (Depo-provera R 250 mg tablets; Hexal)
- Chlamydia trachomatis L2 stock (1 x 107 bacteria: ATCC VR-902 B)
- RPMI Media 1640+ GlutaMax (Gibco, catalog number: 11875)
- Heat inactivated Fetal Calf Serum (FCS) (Sigma, catalog number: F2442)
- DNeasy blood and tissue kit (Qiagen, catalog number: 69504)
- Tissue homogenization beads (MP Biomedicals, catalog number: 6913-500)
- 1x DPBS (Gibco, catalog number: 14190)
- Sucrose (Sigma, catalog number: S0389)
- Glutamic acid (Sigma, catalog number: G1251)
- Nuclease free water (Sigma, catalog number: 7732-18-5)
- SYBR green (Thermo Scientific, catalog number: A25741)
- Primers (Sigma)
- Meglumin diatrizoate (Sigma, CAS: 131-49-7)
- Sodium diatrizoate hydrate (Sigma, CAS: 737-31-5)
- Sodium Citrate dehydrate (Sigma, catalog number: 106447)
- EDTA (Sigma, CAS: 60-00-4)
- HBSS (Sigma, catalog number: H4385)
- Renografin
- 10 mM sodium phosphate (8 mM Na2HPO4 and 2 mM NaH2PO4; Sigma, CAS: 7558-79-4; CAS: 7558-80-7)
- SPG buffer (see Recipes)
- Renografin (see Recipes)
Equipment
- Pipettes
- Incubator (ThermoScientific)
- Centrifuge (Beckman Coulter, model: AvantiTMJ 25I)
- Ultra centrifuge (Beckman Coulter, model: Optima L-80 XP)
- -80 °C freezer (New Brunswick Scientific)
- RT-PCR instrument (ABI 7500)
Software
- Microsoft Excel
- Step One Plus software package (ABI 7500)
- GraphPad Prism 7
Procedure
- Preparation of bacterial stock
Note: The protocol below is for Ct grown in two 75 cm2 flask of cells.
Preparation of chlamydial stock (Mukhopadhyay et al., 2004; Rajeeve et al., 2018).- For infections in mice, Chlamydia trachomatis (Ct) is propagated in mouse embryonic fibroblast (MEFs). Cells are grown in 75 cm2 or 150 cm2 flasks in RPMI with 5% heat inactivated FCS.
- The cells are infected with Ct for 48 h. The cells are scraped with a rubber scraper and collected in a sterile 50 ml Falcon.
- The cells are lysed using glass beads for 3 min by vortexing to release the bacteria.
- The lysate is pelleted to remove the cell debris by centrifuging at 1,344 x g for 10 min at 4 °C.
- The lysate from Step A4 is centrifuged at 30,000 x g for 30 min. The pellet is washed with 10 ml 1x SPG buffer at 30,000 x g for 30 min.
- Now the pellet is resuspended in 2 ml of SPG buffer and passed 5 times through 20 G syringe and later 18 G syringe.
- To prevent any cell debris, the bacterial suspension should be purified using renografin gradient.
- Prepare a step 20-50% (vol/vol) Renografin gradient in sterile 38.0-ml high-speed centrifuge tubes (Beckman Coulter polycarbonate coated).
- The gradient should be made with 3.0 ml of 50% Renografin and 7.0 ml of 20% Renografin carefully layered above the 50% Renografin.
- Add 2.0 ml of the Ct suspension onto the top of the 20% phase.
- Centrifuge at 60,000 x g for 60 min. The EB will appear as a distinct band the 20-50% interface. This layer should be carefully separated and diluted with ice-cold SPG buffer. Repeat a centrifugation step at 30,000 x g and collect the bacterial pellet (it can be very sticky).
- Resuspended bacterial suspension is made into small aliquots (20-50 μl) and frozen at -80 °C until used. One aliquot should be used to check the multiplicity of infection (MOI). We need 1 x 106-1 x 107 bacteria to infect a mouse.
- Determine the multiplicity of infection (MOI)
- Hela cells are plated in duplicate on 12-well plate with glass slides to get a 60-70% confluency next day.
- Different concentrations of bacterial aliquot in different dilution from 1:10 to 1:10-10 are used to infect each well (e.g., 0.25 μl, 0.50 μl, 1 μl, 2 µl and so on).
- After 48 hpi, the MOI is calculated by counting the number of inclusions and number of cells. A MOI of 1 indicates one bacterium infects one cell thereby all cells are infected in a well.
- Selection of mice
Trans cervical infections are carried out in female mouse, which are sexually matured minimum 8 weeks old. Five-ten female mice should be considered per treatment group. Mice in each experiment should be age-matched and cage mates should be randomly distributed into different treatment groups to avoid cage effect. - Transcervical infection
- To establish a genital tract infection, it is necessary to control the shedding from the uterus. Depo-provera, the hormone progesterone prevents ovulation, thus synchronize the estrus cycle and increase susceptibility to Chlamydia infection. Five days before transcervical infection, mice are injected subcutaneously with 2.5 mg of DepoProvera (medroxy-progesterone acetate): Dissolve one tablet (250 mg) in PBS and use 200 μl per mouse.
- On the day of trans cervical infection procedure the bacterial stocks are thawed in ice. One should also carry sterile PBS in 50 ml Falcon, 200 μl pipette and Non-surgical embryo transfer (NSET) device (Figure 2).
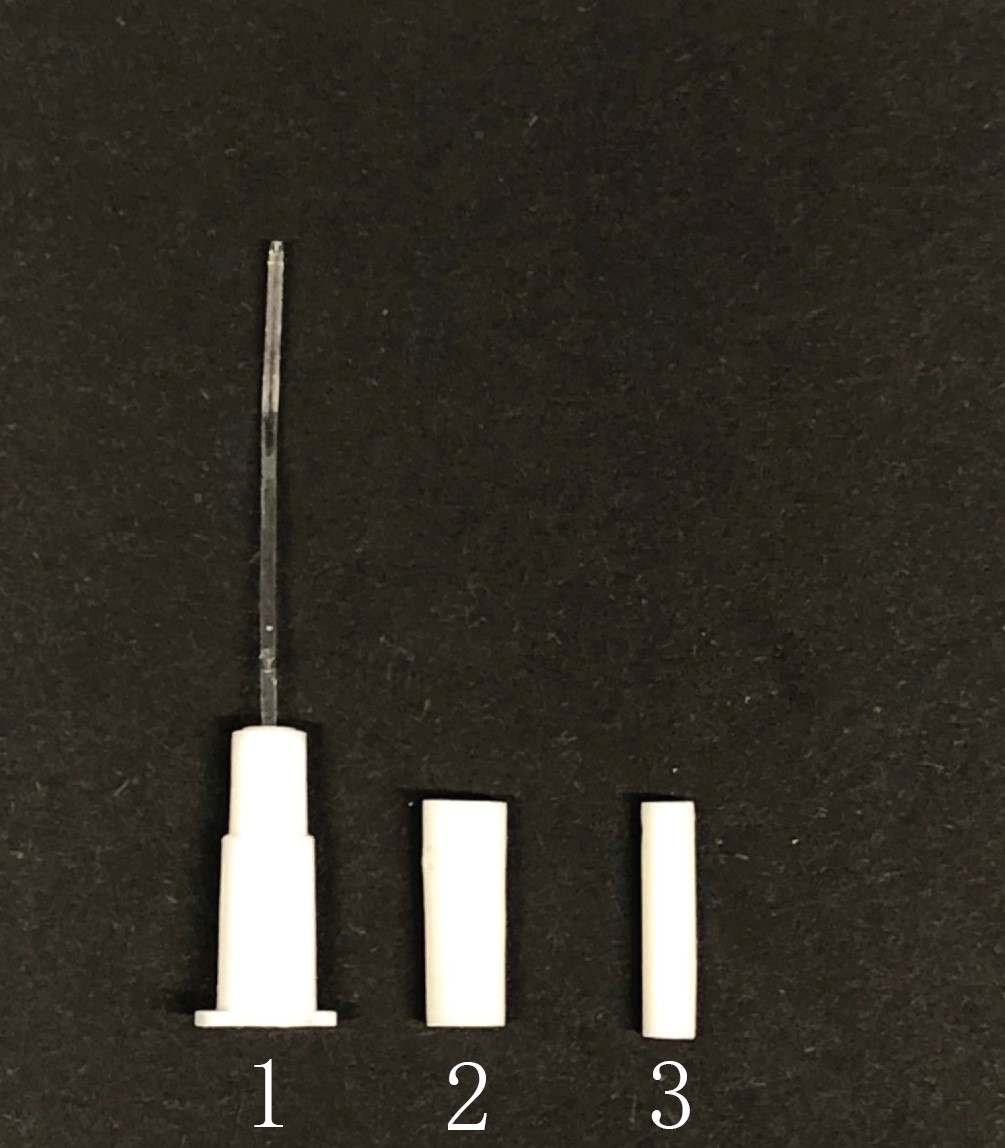
Figure 2. Non-Surgical Embryo Transfer (NSET) device. 1, 2 and 3 indicates the parts of the NSET device. - The infection procedure should be performed in a biosafety level 2 facility. The mouse is retrieved from the cage and placed on the top of the grill. Gently hold the tail and allow the mice to grip on the top of the cage. Now orient the mouse so that the posterior part is visible.
- Gently hold the tail between thumb and forefinger and pull upward that the feet come off the cage. Now use the ring finger to push the back of the mice to angle the hip upwards to easily access the vagina.
- Carefully insert a 200 μl pipette tip into the vagina nearly 0.2 cm deep and aspirate liquid that accumulates the cervix. Make sure not to insert too deep that might hurt the mice and lead to internal injury.
- Now slowly insert the NSET device into the vaginal vault by placing the parts in the order 1, 2 and 3 (Figure 2) as shown in the image and video below (Figure 3) in the order. Pass the needle up and down until it passes through the cervical opening. This needs a few trails and one can use bromophenol blue to confirm passing the cervix (see Video 1).
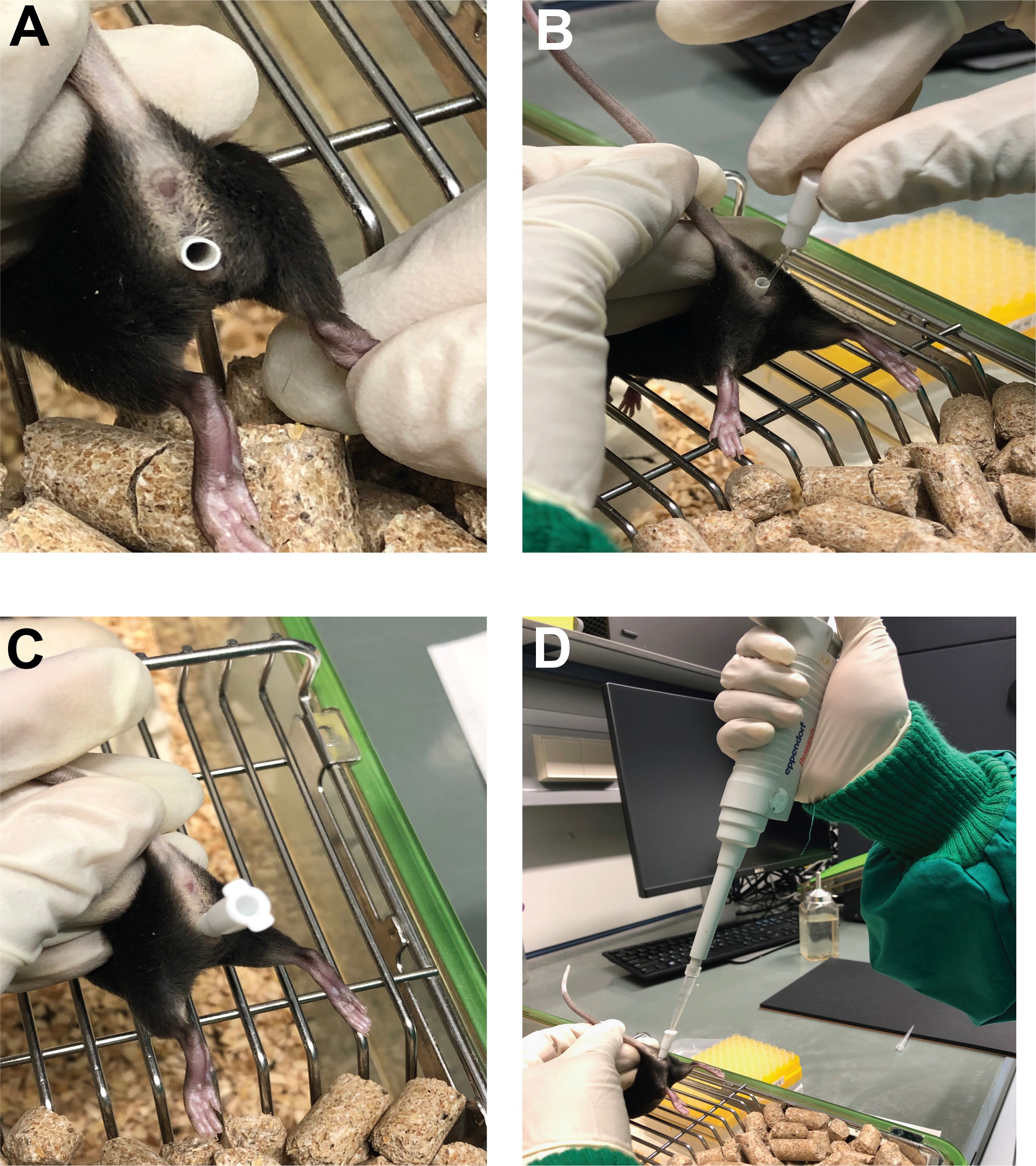
Figure 3. Steps in transcervical infection. A. The mouse is held by the tail as explained above (Step D4) and insert the adaptor of the NSET device into the vagina. B. The NSET needle is further inserted through the adaptor till it reaches the cervix of the mice. C. Make sure that the needle is completely inserted into the cervix. D. One can insert the pipette into the NSET needle and load the bacterial inoculum (refer to the Video 1).Video 1. Steps in transcervical infection. This video was made at the University of Würzburg under the allowance A2 55.5-2531.01-49/12. The animal experiments were performed in accordance with protocols approved by animal care and experimentation of German Animal Protection Law approved under the Animal (Scientific Procedures) Act 1986 (project license 55.2-2532-2-762). - Pipette the required MOI of bacteria through the NSET device and load the bacteria to the uterine fundus. Keep the mouse with the hip upward for 3-5 min to ensure the proper spread of the inoculum. Place the mouse back to its home cage.
- Bacterial load determination
Copy number detection via RT-PCR- The mice were euthanized 7 days post-infection and the uterine horns were taken for further analysis.
- The uterine horns were homogenized in SPG buffer and DNA was isolated using DNeasy blood and tissue kit (Qiagen) as per the manufacturers’ protocol.
- Quantitative PCR was used to enumerate Chlamydia and host genome copy number.
- To determine the copy number a standard curve was done first with a plasmid encoding the chlamydial lytA gene and mouse gene Synectin. One can use any bacterial/host gene for this purpose. The following primers were used for amplifying the C. trachomatis lytA gene that was cloned into the vector: fwd, 5′-TCTAAAGCGTCTGGTGAAAGCT-3′ and rev, 5′-GAAATAGCGTAGTAATAATACCCG-3′. Normalization of bacterial genome to that of the host was performed using mouse synectin primers: fwd, 5′-ACTAATGTCAAGGAGCTGTACG-3′ and rev, 5′-CCTCCGACTTGAACACTTCC-3′.
For creating the standard curve first, the gene of interest (lyt A from Chlamydia/Synectin from mouse) should be cloned in a suitable plasmid for example TOPO pCR 2.1 vector (Life Technologies, Germany). Different dilutions of the Plasmid are used to perform a standard curve. This gives the primary information on the DNA copy number and the corresponding CT value (Figure 4).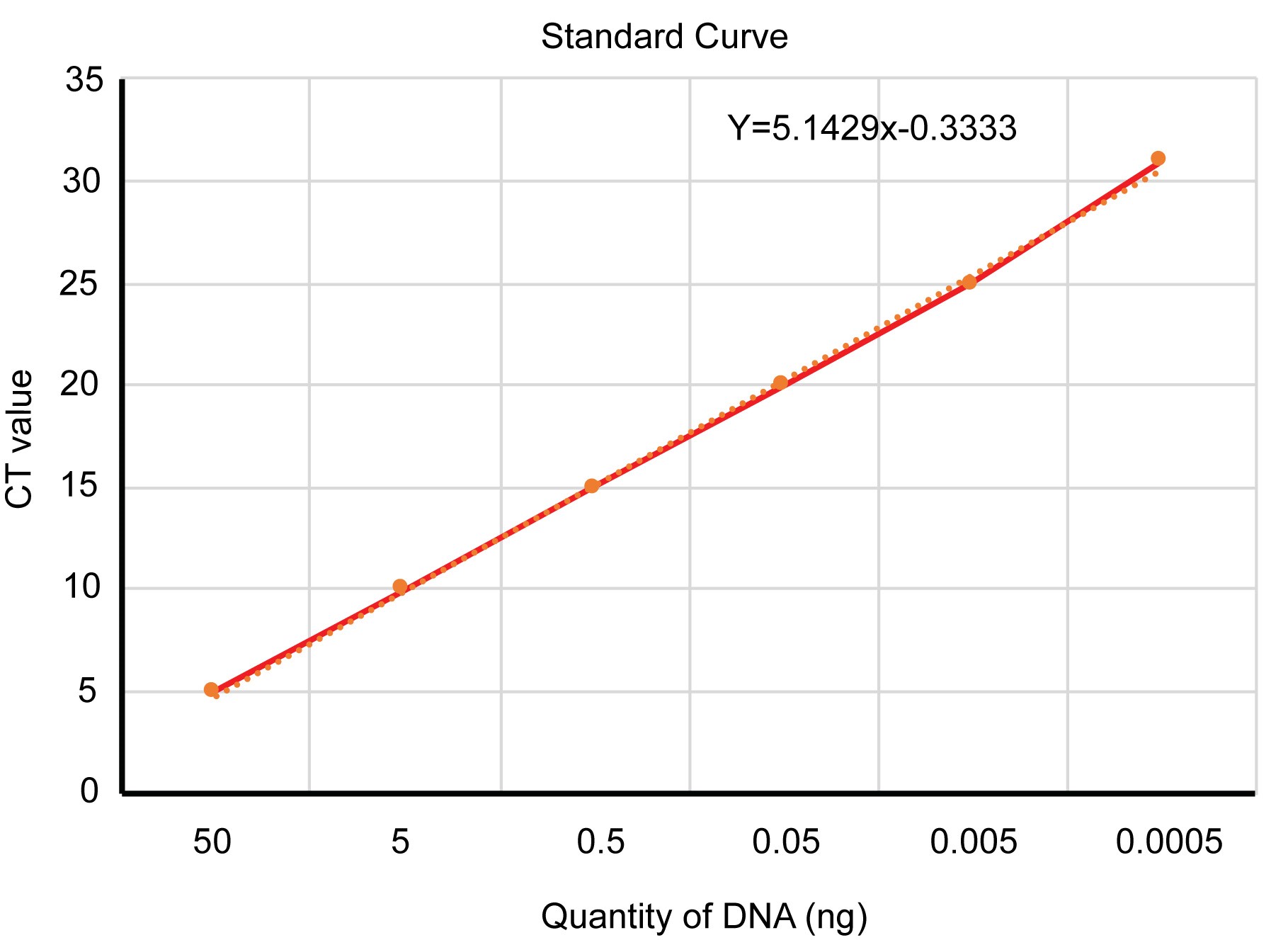
Figure 4. An example of determining the copy number. Different dilutions of plasmid DNA is used to make a standard curve. From the slope of the given line, the amount of DNA present in an unknown sample can be calculated (in this graph the X value) from a corresponding CT value (Y value).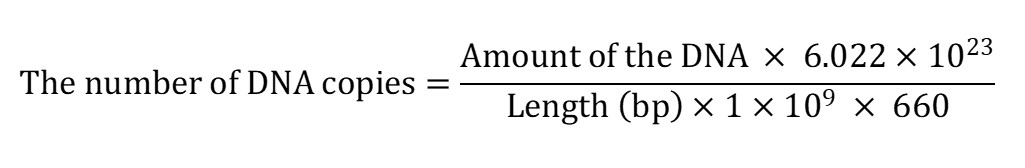
- From the standard curve, the number of copies is determined per µl of the sample as the ratio of chlamydial genome to the host genome.
Data analysis
Data were analyzed using Step One Plus software package (Applied Biosystems) and expressed as the ratio of chlamydial genome to host genome (lytA/synectin). GraphPad Prism 7 was used to generate a scatter column chart and perform statistical analysis. One-way analysis of variance (ANOVA) with Newman–Keuls multiple-comparison tests was performed with the significance level set to less than 0.01.
Alternate method of Copy number detection:
Since the RT-PCR method can detect DNA from dead bacteria, an alternate method of MOI detection from the mouse can also be used.
- HeLa cells are plated on glass slides in a 24-well plate.
- The uterine horns were either homogenized in SPG buffer or vaginal swabs are collected in SPG buffer.
- The sample is further vortexed with glass beads, and the titers of the chlamydial organisms released into the supernatants are determined on HeLa cell monolayers in duplicate.
- The number of inclusions forming units will give the bacterial load.
- The total number of IFU per swab are calculated on the basis of dilution factors, inoculation doses, and total sample volumes. An average is taken from the serially diluted and duplicate samples for any given swab. The calculated total number of IFU per swab are converted into log10, and the log10 IFU counts are used to calculate the mean and standard deviation at each time point.
Notes
- It is important to have the required MOI in 50-100 μl of SPG buffer.
- The bacterial stock that is being used should be tested for mycoplasma contamination.
- Young mice, which are sexually matured, should be selected for the study. Aged mice do not show a tubal pathology and do not serve as a suitable model.
- Include 5-10 mice per condition and also a PBS injected mice, a hormone induced heat inactivated Chlamydia infected mice as control.
- When selecting a gene for copy number analysis, one should take care for a single copy gene, if multiple copy is present then the calculations must be adapted accordingly.
Recipes
- SPG Buffer
10 mM sodium phosphate (8 mM Na2HPO4 and 2 mM NaH2PO4)
220 mM Sucrose
0.5 mM Glutamic acid, pH 7.4 - Renografin
26 g Meglumin diatrizoate
4 g Sodium diatrizoate hydrate
0.16 g Sodium Citrate dehydrate
0.02 g EDTA dissolve in 50 ml HBSS, pH 7.4
Acknowledgments
We acknowledge Heike Czotscher and Nadine Vollmuth for mouse keeping/genotyping and Prof. Thomas Rudel for providing the mouse facility. This study was optimized with the ‘Frauenburo funding for junior scientist’ from DFG for KR and GRK2157 grant from Thomas Rudel. The above protocol is adapted from Rajeeve et al., 2018. KR developed the technique and wrote the manuscript. RS edited the manuscript and provided the photos and videos required for the publication.
Competing interests
The authors declare no competing interest.
Ethics
Statistical analysis was performed to decide the sample size used in mouse infection by the Institute of Mathematics, University of Würzburg under the allowance A2 55.5-2531.01-49/12. All animal experiments were performed in accordance with protocols approved by animal care and experimentation of German Animal Protection Law approved under the Animal (Scientific Procedures) Act 1986 (project license 55.2-2532-2-762).
References
- Abdelrahman, Y. M. and Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol Rev 29(5): 949-959.
- Buckner, L. R., Amedee, A. M., Albritton, H. L., Kozlowski, P. A., Lacour, N., McGowin, C. L., Schust, D. J. and Quayle, A. J. (2016). Chlamydia trachomatis infection of endocervical epithelial cells enhances early HIV transmission events. PLoS One 11(1): e0146663.
- Carmichael, J. R., Tifrea, D., Pal, S. and de la Maza, L. M. (2013). Differences in infectivity and induction of infertility: a comparative study of Chlamydia trachomatis strains in the murine model. Microbes Infect 15(3): 219-229.
- Darville, T., Andrews, C. W., Jr., Laffoon, K. K., Shymasani, W., Kishen, L. R. and Rank, R. G. (1997). Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect Immun 65(8): 3065-3073.
- De Clercq, E., Kalmar, I. and Vanrompay, D. (2013). Animal models for studying female genital tract infection with Chlamydia trachomatis. Infect Immun 81(9): 3060-3067.
- Gondek, D. C., Olive, A. J., Stary, G. and Starnbach, M. N. (2012). CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol 189(5): 2441-2449.
- Morrison, R. P. and Caldwell, H. D. (2002). Immunity to murine chlamydial genital infection. Infect Immun 70(6): 2741-2751.
- Mukhopadhyay, S., Clark, A. P., Sullivan, E. D., Miller, R. D. and Summersgill, J. T. (2004). Detailed protocol for purification of Chlamydia pneumoniae elementary bodies. J Clin Microbiol 42(7): 3288-3290.
- O'Connell, C. M. and Ferone, M. E. (2016). Chlamydia trachomatis genital infections. Microb Cell 3(9): 390-403.
- Pal, S., Tifrea, D. F., Zhong, G. and de la Maza, L. M. (2018). Transcervical inoculation with Chlamydia trachomatis induces infertility in HLA-DR4 transgenic and wild-type mice. Infect Immun 86(1).
- Poston, T. B., Gottlieb, S. L. and Darville, T. (2017). Status of vaccine research and development of vaccines for Chlamydia trachomatis infection. Vaccine 37(50): 7289-7294.
- Rajeeve, K., Das, S., Prusty, B. K. and Rudel, T. (2018). Chlamydia trachomatis paralyses neutrophils to evade the host innate immune response. Nat Microbiol 3(7): 824-835.
- Ramsey, K. H., DeWolfe, J. L. and Salyer, R. D. (2000). Disease outcome subsequent to primary and secondary urogenital infection with murine or human biovars of Chlamydia trachomatis. Infect Immun 68(12): 7186-7189.
- Shah, A. A., Schripsema, J. H., Imtiaz, M. T., Sigar, I. M., Kasimos, J., Matos, P. G., Inouye, S. and Ramsey, K. H. (2005). Histopathologic changes related to fibrotic oviduct occlusion after genital tract infection of mice with Chlamydia muridarum. Sex Transm Dis 32(1): 49-56.
- Starnbach, M. N. (2018). Action needed on Chlamydia Vaccines. Trends Microbiol 26(8): 639-640.
- Sturdevant, G. L. and Caldwell, H. D. (2014). Innate immunity is sufficient for the clearance of Chlamydia trachomatis from the female mouse genital tract. Pathog Dis 72(1): 70-73.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rajeeve, K. and Sivadasan, R. (2020). Transcervical Mouse Infections with Chlamydia trachomatis and Determination of Bacterial Burden. Bio-protocol 10(3): e3506. DOI: 10.21769/BioProtoc.3506.
Category
Microbiology > Microbe-host interactions > Bacterium
Immunology > Animal model > Mouse
Molecular Biology > DNA > DNA quantification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











