- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Steady-state and Flux-based Trehalose Estimation as an Indicator of Carbon Flow from Gluconeogenesis or Glycolysis
Published: Vol 10, Iss 1, Jan 5, 2020 DOI: 10.21769/BioProtoc.3483 Views: 5537
Reviewed by: Valerian DORMOYAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Intra- and Extracellular Glucose in Mycelium of Fusarium oxysporum
Carmen Ruiz-Roldan and M. Isabel G. Roncero
Jul 20, 2016 10900 Views

Observing Nutrient Gradients, Gene Expression and Growth Variation Using the "Yeast Machine" Microfluidic Device
Zoran S. Marinkovic [...] Pascal Hersen
Jul 5, 2020 5114 Views
Abstract
Trehalose (and glycogen) is a major storage carbohydrate in many cells, including S. cerevisiae. Typically, trehalose (a disaccharide of glucose) is synthesized and stored through gluconeogenesis. However, trehalose can also be made directly from glucose, if glucose-6-phosphate is channeled away from glycolysis or pentose phosphate pathway. Therefore, analyzing trehalose synthesis, utilization or its accumulation, can be used as a sentinel read-out for either gluconeogenesis or rewired glucose utilization. However, the steady-state measurements alone of trehalose cannot unambiguously distinguish the nature of carbon flux in a system. Here, we first summarize simple steady-state enzymatic assays to measure trehalose (and glycogen), that will have very wide uses. Subsequently, we describe methods of highly sensitive, quantitative LC-MS/MS based to measure trehalose. We include methods of 13C stable-isotope based pulse-labeling experiments (using different carbon sources) with which to measure rates of trehalose synthesis, from different carbon metabolism pathways. This approach can be used to unambiguously determine the extent of carbon flux into trehalose coming from gluconeogenesis, or directly from glucose/glycolysis. These protocols collectively enable comprehensive steady-state as well as carbon flux based measurements of trehalose. This permits a dissection of carbon flux to distinguish between cells in a gluconeogenic state (conventionally leading to trehalose synthesis), or cells with rewired glucose metabolism (also leading to trehalose synthesis). While the methods presented are optimized for yeast, these methods can be easily adapted to several types of cells, including many microbes.
Keywords: TrehaloseBackground
Carbon flux estimates are excellent indicators of overall cellular metabolic states (Metallo and Vander Heiden, 2013; Antoniewicz, 2018; Jang et al., 2018). Typically, glucose consumption towards high glycolysis and the pentose phosphate pathway reflect a ‘growth’ metabolic state, and will lead to high nucleotide synthesis and energy production. Alternately, high rates of gluconeogenesis from alternate carbon sources, which lead to the formation of the storage carbohydrates trehalose and glycogen, indicate a ‘starvation-like’ metabolic state. To easily identify cell states, estimates of carbon flux into trehalose, when properly used, can distinguish between high gluconeogenic states or simply rewired glucose metabolism due to other causes, as described below.
Trehalose has been best studied as a major storage sugar in a range of organisms, ranging from yeast, nematodes, flies, plants and more (Elbein et al., 2003; Iturriaga et al., 2009). Functionally, trehalose has multiple roles in survival and desiccation tolerance (Lillie and Pringle, 1980; Behm, 1997; Erkut et al., 2011; Iordachescu and Imai, 2011; Tapia and Koshland, 2014). Also trehalose consumption enables cells to exit from quiescence (Shi et al., 2010; Dhawan and Laxman, 2015; Laporte et al., 2017). High amounts of trehalose accumulation in cells is a metabolic signature for cells in a ‘quiescence-like’ state (Gray et al., 2004; Erkut et al., 2011; De Virgilio, 2012). Conventionally, trehalose is primarily made in cells during glucose limitation, when there is very high gluconeogenic flux from alternate carbon sources. This is illustrated in Figure 1. However, trehalose is also produced during glycolysis when glucose is consumed in a constant ‘futile’ cycle in order to balance glycolysis (Van Heerden et al., 2014) (Figure 1). In this case, trehalose does not accumulate, and glucose-6-phosphate is largely channeled into the pentose phosphate pathway (PPP) and glycolysis. However, if a rewiring of either the PPP and/or the lower arm of glycolysis occurs, this can result in greater flux into trehalose synthesis and accumulation (Figure 1). This second mode of trehalose synthesis, which can occur due to different (and sometimes distant) causes, all of which will reflect a ‘starvation-state’, is far less appreciated. In recent studies, we observed such a rewiring of carbon metabolism in specific tRNA modification mutants, resulting in such a ‘starvation-state’ (Gupta et al., 2019). Notably, carbon flux from glucose into trehalose was a key read-out used to establish and characterize these metabolic states (Gupta et al., 2019). In this context, a limitation in free (inorganic) phosphate controlled this metabolic rewiring. Here, glucose-6-phosphate (from glucose) was routed towards trehalose synthesis, since trehalose synthesis also releases phosphates (Gupta et al., 2019).

Figure 1. Trehalose synthesis is made either from glucose during glycolysis (in a futile cycle), or as an end product of gluconeogenesis. The green arrows indicate gluconeogenic flux (which can come from oxaloacetate (OAA), aspartate or other gluconeogenic carbon sources), while the red arrows indicate carbon flux during glycolysis (coming from glucose).
Using the protocols described in this manuscript to measure steady-state trehalose concentrations, we had noted that such an approach was insufficient to establish a gluconeogenic state (Gupta et al., 2019). However, by using different 13C stable-isotope based approaches (described below), we could measure flux into trehalose, and determine whether trehalose is being made via to gluconeogenesis, or from altered glycolysis (Figure 1). Here, by using labeled glucose, we can determine carbon flux into trehalose directly from glucose (Gupta et al., 2019). Using alternate 13C-labeled carbon sources (primarily aspartate), we can unambiguously estimate trehalose synthesized due to gluconeogenesis (or alternately distinguish this from trehalose made directly from glucose). This combined use of steady-state and 13C-labeled carbon-flux measurements of trehalose is therefore extremely powerful as an analytical approach. This can quantitatively and qualitatively distinguish metabolic states of cells, and indicate the nature of the carbon flux outputs. While the protocols described here were optimized in yeast, they are easily adaptable to almost any cellular system (of suspension or adherent cells), and so have broad uses, particularly in microbial systems. We also note that this approach towards estimating only flux towards trehalose is complementary to, and also simpler and more comprehensive than other methods for analysis of carbon flux, described in other studies (Walvekar et al., 2018; Gupta et al., 2019). We collectively present a range of methods to unambiguously, quantitatively estimate trehalose amounts, as well as carbon flux into trehalose, all of which can be used to establish overall carbon metabolism and metabolic states in cells.
Materials and Reagents
- For enzymatically estimating steady-state trehalose and glycogen amounts
- 15 ml Falcon tubes (Tarsons, catalog number: 546021)
- 50 ml Falcon tubes (Tarsons, catalog number: 546041)
- 1.5 ml screw cap tubes (Genetix, catalog number: 3744.S.X-A)
- 1.5 ml Eppendorf tubes (Tarsons, catalog number: 500010)
- 96-well plate (Greiner Bio-One, catalog number: 650180)
- Porcine trehalase (Sigma-Aldrich, catalog number: T8778)
- Aspergillus niger amyloglucosidase (Sigma-Aldrich, catalog number:10115)
- Glucose estimation kit (Sigma-Aldrich, catalog number: GAGO20)
- Sodium carbonate (Fisher Scientific, catalog number: 15955)
- Acetic acid (Fisher Scientific, catalog number: 11005)
- Sodium acetate (Sigma-Aldrich, catalog number: W302406)
- Sulfuric acid (Fisher Scientific, catalog number: 29995)
- 0.25 M sodium carbonate (50 ml) (see Recipes)
- 1 M glacial acetic acid (50 ml) (see Recipes)
- 0.2 M sodium acetate, pH 5.2 (50 ml) (see Recipes)
- 12 N sulfuric acid (25 ml) (see Recipes)
- Rich medium (see Recipes)
- Synthetic minimal medium (see Recipes)
- For MS/MS based trehalose measurements
- 50 ml Falcon tubes (Tarsons, catalog number: 546041)
- 1.5 ml Eppendorf tubes (Tarsons, catalog number: 500010)
- Reverse-phase HPLC column (recommended column: Synergi 4 µm Fusion-RP 80A from Phenomenex, catalog number: 00D-4424-E0)
- HPLC grade methanol (Fisher Scientific, catalog number: A456-4)
- HPLC grade water (Fisher Scientific, catalog number: W6-4)
- [U-13C6] glucose (Cambridge Isotope Laboratories, catalog number: CLM-1396-PK)
- [U-13C4] aspartate (Cambridge Isotope Laboratories, catalog number: CLM-1801-H-PK)
- Trehalose (Sigma-Aldrich, catalog number: T951)
- 100% acetonitrile (Fisher Scientific, A955-4)
- Ammonium acetate (Fisher Scientific, catalog number: 17836-50G)
- Ethanol (Millipore Sigma, catalog number: 1009830511)
- 25% acetonitrile with 5 mM ammonium acetate (see Recipes)
- 60% methanol (100 ml) (see Recipes)
- 75% ethanol (10 ml) (see Recipes)
Equipment
- For enzymatically estimating trehalose and glycogen amounts
- Shaker incubator for growing cells (Thermo Scientific, model: MaxQ 6000)
- Cold centrifuge for 50/15 ml Falcon tubes (Eppendorf, model: 5702R)
- Cold high-speed bench top centrifuge for 1.5 ml Eppendorf tubes (Eppendorf, model: 5424R)
- Dry heating block or water incubator (at 98 °C, 37 °C) (IKA, model: Dry Block Heater1)
- Hybridization oven (or rotatory incubator) (at 37 °C, 57 °C) (Thermo Fisher Scientific, model: 6240)
- Plate reader (Molecular Devices, model: SpectraMax M series Multi-mode microplate Reader)
- For efficient total metabolite extractions from cells
- Shaker incubator for growing cells (Thermo Scientific, model: MaxQ 6000)
- Rocking platform
- 60% methanol dewar (set at -45 °C)
- Cold centrifuge for 50 ml Falcon tubes (set at -5 °C) (Eppendorf, model: 5702R)
- Cold high-speed bench top centrifuge for 1.5 ml Eppendorf tubes (set at -5 °C) (Eppendorf, model: 5424R)
- Vortexer mixer (IKA, model: vortex Genius 3)
- Dry heating block or water incubator (at 80 °C) (IKA, model: Dry Block Heater1). Water incubator/bath is better to use
- Room temperature high-speed bench top centrifuge for 1.5 ml Eppendorf tubes (Eppendorf, model: 5418)
- Vacuum dryer (Eppendorf Concentrator plus, model: 5305000304)
- HPLC and mass spectrometry
- Agilent’s 1290 infinity series UHPLC system
- ABSCIEX QTRAP 6500 (in triple-quadrupole mode) or a Thermo Scientific TSQ Vantage Triple Stage Quadrupole Mass Spectrometer
HPLC requirements: any HPLC system coupled to an appropriate mass spectrometer would be fine. In our experiments, we have used Agilent’s 1290 infinity series UHPLC system. Here, reverse phase high performance liquid chromatography (HPLC) is performed where Synergi 4 µm Fusion-RP 80A column acts as the stationary phase and gradient of 5 mM ammonium acetate in water and 100% acetonitrile acts as the mobile phase.
Mass spectrometry requirements: Since the trehalose measurements are quantitative, and a broad range of concentrations of trehalose need to be measured, the mass spectrometer used must be capable of accurate measurements over a broad linear dynamic range. For this, a standard triple-quadrupole type mass spectrometer used in MS/MS mode is recommended, however suitable Orbitrap or Q-TOF (triple quadrupole) mass spectrometers may also be used. In our studies, we have used an ABSCIEX QTRAP 6500 (in triple-quadrupole mode) or a Thermo Scientific TSQ Vantage Triple Stage Quadrupole Mass Spectrometer.
Software
- Mass spectrometry data analysis software: Thermo Xcalibur software (Qual and Quan browsers) and AB SCIEX MultiQuant software 3.0.1
- Graphing/plotting software: Microsoft Excel, GraphPad Prism 7 or any other suitable graphing program or R-package
Procedure
- Enzymatic measurement of trehalose and glycogen to estimate steady-state amounts
- For trehalose and glycogen measurements, grow the cells in appropriate medium at 30 °C. The growth medium can be either rich medium (Recipe 8) or synthetic minimal medium (Recipe 9).
- Harvest equal number of cells for different samples (~10 A600 for yeast cells, as measured using a spectrophotometer, for each estimation of trehalose and glycogen) by transferring the cells to either 15- or 50-ml Falcon tube, centrifuge the tubes quickly at 3,000 x g for 3 min.
- Carefully remove the supernatant and wash the cells once with 1 ml of ice-cold water in a 1.5 ml screw cap tube by centrifugation at maximum speed for 15 s.
- Carefully remove the supernatant using either the pipette tip or a syringe connected to vacuum.
Note: This step is important for the removal of residual glucose from the medium. After this step, the cells can be frozen in liquid nitrogen and stored at -80 °C until further use. - Lyse the cells by resuspending the cell pellet in 0.25 ml of 0.25 M sodium carbonate and incubate the tube at 95-98 °C for 4 h.
Note: The sodium carbonate helps in alkali-based permeabilization of the cells, which allows trehalase and amyloglucosidase enzymes to digest trehalose and glycogen respectively. This step can be done either in the dry heating block or water incubator with occasional mixing of the tubes. - Add 0.15 ml of 1 M glacial acetic acid and 0.6 ml of 0.2 M sodium acetate to the tube to adjust the pH of the solution to 5.2.
Note: Glacial acetic acid and sodium acetate are added to bring the pH of the cell suspension to an acidic range, since the optimum pH for the trehalase and amyloglucosidase enzymes are ~5.7 and ~5 respectively. Do not mix too vigorously at this step, as there is production of carbon dioxide. - For trehalose and glycogen measurements, add 0.05 U/ml of trehalase enzyme and 1-2 U/ml of amyloglucosidase enzymes respectively. Incubate the tubes overnight with constant rotation in the hybridization oven (or rotatory incubator) at 37 °C for digestion of trehalose and at 57 °C for digestion of glycogen.
- Centrifuge the tubes at maximum speed, room temperature for 3 min. Carefully transfer the supernatant to a fresh 1.5 ml Eppendorf tube without disturbing the pellet.
- The assay for the glucose released from these digestions using a Glucose (GO) Assay Kit is as per the manufacturer’s protocol. Briefly, transfer either undiluted sample (0.04 ml) or suitably diluted sample (1:1 or 1:4) in triplicates to a 96-well plate. Add 0.08 ml of reconstituted glucose oxidase/peroxidase/o-dianisidine reagent to each well. Incubate the plate at 37 °C for 30 min. Stop the reaction by adding 0.08 ml of 12 N sulfuric acid and read the absorbance at 540 nm in a plate reader. The concentration of released glucose (mg/ml) is determined using the standard curve.
- Preparing metabolite extracts (to extract trehalose and other metabolites from cells)
Note: This is shown as the schematic in Figure 2.- Grow the cells in appropriate medium at 30 °C. The growth medium can be either rich medium (Recipe 8) or synthetic minimal medium (Recipe 9). Harvest an equal number of cells for different samples (~6-10 A600 for yeast cells using a spectrophotometer) during the logarithmic phase of growth. The volume of the culture to be harvested can range from 5 to 10 ml.
- Quench metabolism in the cells for 5 min in 4 volumes of 60% methanol in a 50 ml Falcon tube (maintained at -40 °C to -45 °C).
Note: Use dry ice to maintain the temperature of the 60% methanol contained in the dewar at -40 °C to -45 °C. Do not cool to below -50 °C as cells will freeze at that temperature. In case, the temperature drops below -50 °C, bring it to -45 °C by pouring 60% methanol maintained at room temperature into the dewar. Once the temperature is set at -40 °C to -45 °C, it is maintained for a while, and therefore stringent temperature controlling methods are not needed at this step. - Centrifuge the tubes at 1,000 x g for 3 min (set at -5 °C).
Note: Cool all the centrifuges to -5 °C even before harvesting the cells. - Carefully remove the supernatant and then resuspend the pellet in cold 0.7 ml of 60% methanol (maintained at -45 °C).
- Transfer the suspension to 1.5 ml Eppendorf tubes and centrifuge this suspension at 1,000 x g for 3 min (set at -5 °C).
Note: Keep the 1.5 ml Eppendorf tubes chilled by placing them in 60% methanol dewar (set at -45 °C). - Carefully remove the supernatant using a pipette (and not vacuum based aspiration), and resuspend the pellet in 1 ml of 75% ethanol.

Figure 2. Steady-state measurements of trehalose using (A) either an enzyme based colorimetric assay or (B) HPLC-MS/MS based analysis. The enzymatic assay is simple and low-cost, and measures total trehalose. The LC-MS/MS based analysis is very sensitive (100-1000 fold higher than the enzyme based assay), and can unambiguously detect and quantify total trehalose in the extracts, and can also be used to differentiate the source of trehalose when used in conjunction with stable-isotope labeling. - Keep the tubes at 80 °C for 3 min, with intermittent vortexing, followed by incubation on ice for 5 min.
- Centrifuge the tubes at maximum speed for 1 min at room temperature to pellet the extracted cells.
- Transfer 0.96 ml of the supernatant to a fresh 1.5 ml Eppendorf tube, centrifuge again at maximum speed for 10 min at room temperature to pellet any remaining cell debris.
- Transfer 0.45 ml of the supernatant to two fresh 1.5 ml Eppendorf tubes.
- Dry the supernatant in a vacuum dryer for 3-4 h and then store at -80 °C until further use.
- Estimating trehalose by LC-MS/MS
Note: Also see schematic in Figure 2.- Prepare a stock of 0.25-1 µg/ml trehalose solution to be used as a standard in LC-MS/MS. This solution is prepared such that the effective standard concentration used in each LC-MS/MS run is 5-10 ng. Make multiple dilutions of this stock to determine the linear range for trehalose estimation, such that 1-50 ng of trehalose is loaded onto the column.
- Use appropriate volumes of the solvent solution (Recipe 7) to dissolve the samples. The solvent volume can range from 0.05 ml to 0.5 ml, depending on the concentration of trehalose in the samples and the linear dynamic range of the mass spectrometer.
Note: It will be appropriate to dissolve the sample in smaller volumes at first, and then dilute it further if needed. - Resolve metabolite extracts by injecting into the HPLC column, and analyze the metabolites using MS/MS. Measure trehalose using a negative polarity mode in the mass spectrometer. For further details on the buffers, column parameters, flow rate and the gradient, refer to the method described in detail in Walvekar et al., 2018.
- 13C-labeling experiments for flux based trehalose synthesis estimations
Note: See Figure 3 for a typical labeling experiment workflow.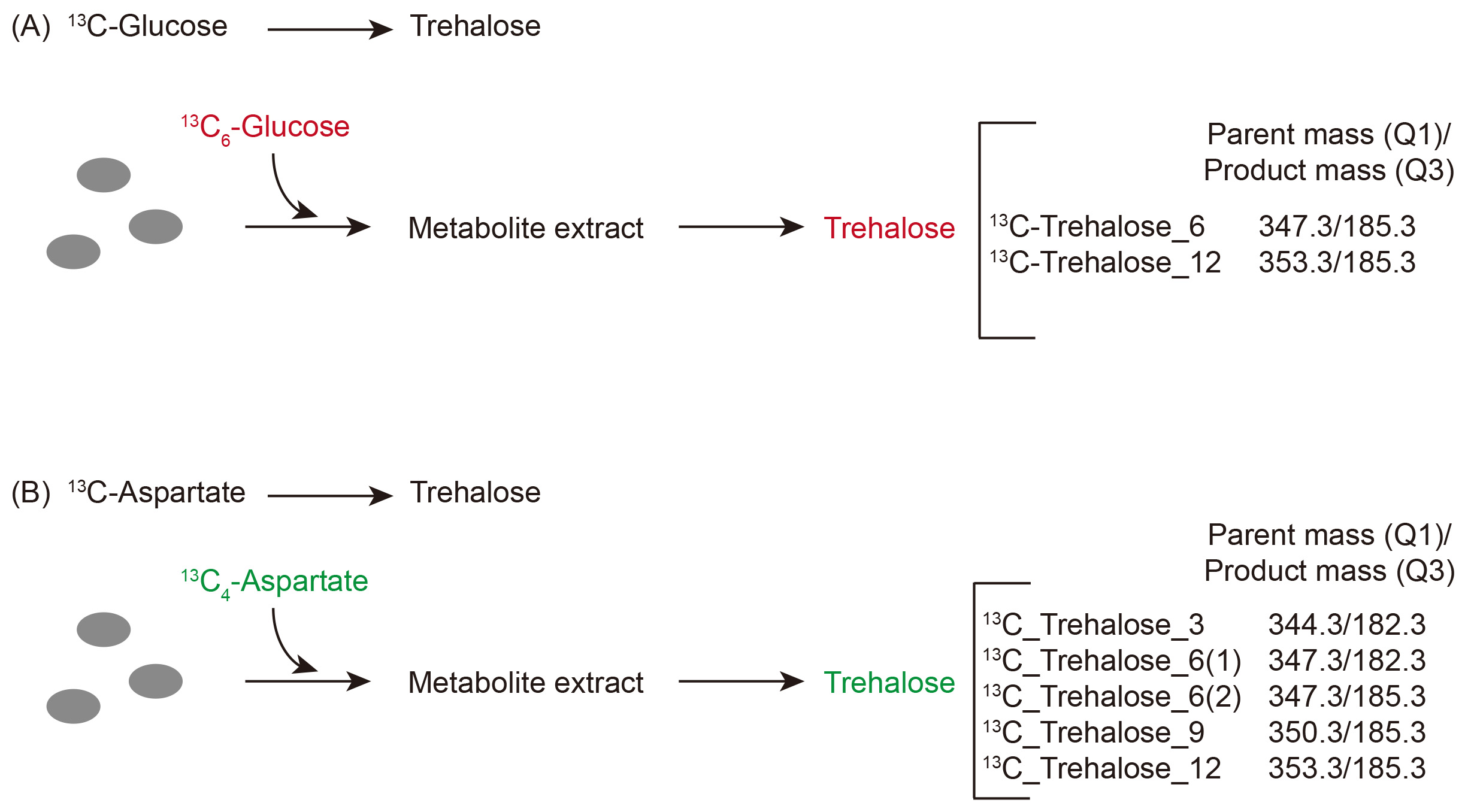
Figure 3. Workflow for a typical carbon-label based estimation of trehalose synthesis. A. A schematic to show the use of stable-isotope labeled glucose to estimate trehalose synthesis coming from glycolysis (i.e., directly from glucose). B. A schematic to show the use of stable-isotope labeled aspartic acid to estimate trehalose synthesis from gluconeogenesis.
13C-glucose labeling (direct incorporation of glucose into trehalose)
- For experiments to estimate the 13C-label incorporation into trehalose from glucose, grow the cells in appropriate medium at 30 °C till the logarithmic phase. Growth medium can be either rich medium (Recipe 8) or synthetic minimal medium (Recipe 9).
- Add labeled [U-13C6] glucose to the culture to reach a ratio of 50% unlabeled to 50% fully labeled glucose for ~5 min or less on a rocking platform (For cells grown in 2% unlabeled glucose, add 2% labeled glucose).
Note: Since labeled glucose gets incorporated into trehalose directly, the incubation time with labeled glucose is critical and should be reduced to the minimum possible to avoid saturation of label incorporation into trehalose. This time should be optimized carefully based on preliminary label incorporation measurements. - Harvest equal numbers of cells for different samples (~6-10 ml of yeast cells in culture at an A600 of ~1) and proceed as described in Step B2.
Note: During harvesting an equal number of cells, measure the A600 of the culture before spiking in labeled glucose to minimize the delay in time.
13C-aspartate labeling (gluconeogenic flux into trehalose)
- For the 13C-label incorporation in trehalose from aspartate, grow the cells in an appropriate medium (containing glycerol/ethanol as carbon source) and collect cells when still in the logarithmic phase.
Note: Alternatively, cells can be either grown in glucose till late-logarithmic phase (post-diauxic), or low glucose concentrations can also be used, depending on the needs of the experiment. - Add labeled [U-13C4] aspartate to a final concentration of 1 mM and incubate for appropriate time.
Note: The time of incubation with labeled aspartate can be optimized based on the linearity of the label incorporation into trehalose. ~15 min can be used as an earliest time-point since there are multiple steps involved in trehalose synthesis from aspartate via gluconeogenesis. - Harvest an equal number of cells for different samples (~6-10 ml of yeast cells at A600 ~1) and proceed as described in Step B2.
Normalizing MS/MS data
We recommend using any two of the three approaches listed below, to ensure proper normalization of data coming from different samples. First, prepare all metabolite extracts from equal cell numbers, inject the same volumes into the mass spectrometer, and carry out all runs together. Second, the use of an internal standard added during extractions is strongly recommended. For this, the use of an unnatural disaccharide, or labeled disaccharide (e.g., melibiose) is ideal. Third, estimates of total ion count (for each injection) can be used to normalize the total amounts of samples injected.
Data analysis
- The area under each metabolite peak is calculated using either appropriate MS software (for example, the Thermo Xcalibur software (Qual and Quan browsers) or AB SCIEX MultiQuant software 3.0.1).
- For estimating steady-state levels of trehalose, either total peak area or relative peak area can be calculated and plotted for different samples.
- For estimating trehalose synthesis by flux experiments, either relative label incorporation or percent label incorporation can be calculated and plotted for all the mass isotopomers of trehalose.
Note: For comparing samples, it is easiest to use a relative normalization, i.e., compare amounts of label incorporated in each sample, in relation to any one sample. Here, peak areas in each sample can be divided by the peak area in the control sample. Subsequent visualizations are done by plotting relative changes, using any presentation style of choice.
Recipes
- 0.25 M sodium carbonate (50 ml)
- Weigh 1.32 g of the powder and dissolve in 50 ml sterile water
- Filter-sterilize the solution
- Store at room temperature (shelf life: several months. Note that Contamination is rare but possible)
- 1 M glacial acetic acid (50 ml)
- To 47.13 ml sterile water, add 2.87 ml of glacial acetic acid
- Store at room temperature for several months
- 0.2 M sodium acetate, pH 5.2 (50 ml)
- Weigh 0.82 g of the powder and dissolve in 50 ml sterile water
- Adjust the pH and filter sterilize the solution
- Store at room temperature (shelf life: several months. Note that contamination is rare but possible.)
- 12 N sulfuric acid (25 ml)
- To 16.7 ml sterile water, add 8.33 ml of sulfuric acid
- Store at room temperature (shelf life: several months)
- 60% methanol (100 ml)
Make it fresh and bring it -45 °C before useWater 40 ml Methanol 60 ml - 75% ethanol (10 ml)
Make it fresh and store at room temperatureWater 2.5 ml Ethanol 7.5 ml - 25% acetonitrile with 5 mM ammonium acetate (100 ml)
Filter sterilize the solutionAcetonitrile 25 ml Ammonium acetate 38.5 mg Water 75 ml
Make it fresh for each experiment, and store at room temperature - Rich medium
1% yeast extract
2% peptone
2% glucose - Synthetic minimal medium
0.67% yeast nitrogen base
2% glucose
with or without amino acids
Acknowledgments
This work was supported in part by a Wellcome Trust–DBT India Alliance intermediate fellowship (IA/I/14/2/501523) to SL, institutional support from inStem, and a Dept. of Science and Technology SERB-NPDF (PDF/2016/000416) to RG. The authors acknowledge the use of the HPLC and mass spectrometry resources in the NCBS/inStem/C-CAMP mass spectrometry facilities.
Competing interests
The authors have no competing interests to declare.
References
- Antoniewicz, M. R. (2018). A guide to 13C metabolic flux analysis for the cancer biologist. Exp Mol Med 50(4): 19.
- Behm, C. A. (1997). The role of trehalose in the physiology of nematodes. Int J Parasitol 27(2): 215-229.
- De Virgilio, C. (2012). The essence of yeast quiescence. FEMS Microbiol Rev 36(2): 306-339.
- Dhawan, J. and Laxman, S. (2015). Decoding the stem cell quiescence cycle--lessons from yeast for regenerative biology. J Cell Sci 128(24): 4467-4474.
- Elbein, A. D., Pan, Y. T., Pastuszak, I. and Carroll, D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13(4): 17R-27R.
- Erkut, C., Penkov, S., Khesbak, H., Vorkel, D., Verbavatz, J. M., Fahmy, K. and Kurzchalia, T. V. (2011). Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21(15): 1331-1336.
- Gray, J. V., Petsko, G. A., Johnston, G. C., Ringe, D., Singer, R. A. and Werner-Washburne, M. (2004). "Sleeping beauty": quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 68(2): 187-206.
- Gupta, R., Walvekar, A. S., Liang, S., Rashida, Z., Shah, P. and Laxman, S. (2019). A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife 8: 44795.
- Iordachescu, M. and Imai, R. (2011). Trehalose and abiotic stress in biological systems. In: Abiotic Stress in Plants-Mechanisms and Adaptations. In: Shanker, A. and Venkateswarlu, B. (Eds.). DOI: 10.5772/22208.
- Iturriaga, G., Suárez, R. and Nova-Franco, B. (2009). Trehalose metabolism: from osmoprotection to signaling. Int J Mol Sci 10(9): 3793-3810.
- Jang, C., Chen, L. and Rabinowitz, J. D. (2018). Metabolomics and isotope tracing. Cell 173(4): 822-837.
- Laporte, D., Jimenez, L., Gouleme, L. and Sagot, I. (2017). Yeast quiescence exit swiftness is influenced by cell volume and chronological age. Microb Cell 5(2): 104-111.
- Lillie, S. H. and Pringle, J. R. (1980). Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143(3): 1384-1394.
- Metallo, C. M. and Vander Heiden, M. G. (2013). Understanding metabolic regulation and its influence on cell physiology. Mol Cell 49(3): 388-398.
- Shi, L., Sutter, B. M., Ye, X. and Tu, B. P. (2010). Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell 21(12): 1982-1990.
- Tapia, H. and Koshland, D. E. (2014). Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol 24(23): 2758-2766.
- van Heerden, J. H., Wortel, M. T., Bruggeman, F. J., Heijnen, J. J., Bollen, Y. J., Planque, R., Hulshof, J., O'Toole, T. G., Wahl, S. A. and Teusink, B. (2014). Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343(6174): 1245114.
- Walvekar, A., Rashida, Z., Maddali, H. and Laxman, S. (2018). A versatile LC-MS/MS approach for comprehensive, quantitative analysis of central metabolic pathways. Wellcome Open Res 3: 122.
Article Information
Copyright
Gupta and Laxman. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Gupta, R. and Laxman, S. (2020). Steady-state and Flux-based Trehalose Estimation as an Indicator of Carbon Flow from Gluconeogenesis or Glycolysis. Bio-protocol 10(1): e3483. DOI: 10.21769/BioProtoc.3483.
- Gupta, R., Walvekar, A. S., Liang, S., Rashida, Z., Shah, P. and Laxman, S. (2019). A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife 8: 44795.
Category
Cell Biology > Cell metabolism > Carbohydrate
Biochemistry > Carbohydrate > Glucose
Microbiology > Microbial metabolism > Carbohydrate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










