- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Random Migration of Cancer Cells in 3D
Published: Vol 10, Iss 1, Jan 5, 2020 DOI: 10.21769/BioProtoc.3482 Views: 6597
Reviewed by: Pilar VillacampaGuillermo Gomezsujan kumar mondal

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Spherical Invasion Assay: A Novel Method to Measure Invasion of Cancer Cells
Stephen D. Richbart [...] Piyali Dasgupta
Feb 20, 2022 5413 Views

Image-based Quantification of Macropinocytosis Using Dextran Uptake into Cultured Cells
Anh H. Le and Laura M. Machesky
Apr 5, 2022 4185 Views

An Experimental Protocol for the Boyden Chamber Invasion Assay With Absorbance Readout
Kathleen C. Brown [...] Piyali Dasgupta
Aug 5, 2024 2761 Views
Abstract
The ability of cancer cells to migrate through a complex three-dimensional (3D) environment is a hallmark event of cancer metastasis. Therefore, an in vitro migration assay to evaluate cancer cell migration in a 3D setting is valuable to examine cancer progression. Here, we describe such a simple migration assay in a 3D collagen-fibronectin gel for observing cell morphology and comparing the migration abilities of cancer cells. We describe below how to prepare the collagen-fibronectin gel castings, how to set up time-lapse recording, how to draw single-cell trajectories from movies and extract key parameters that characterize cell motility, such as cell speed, directionality, mean square displacement, and directional persistence. In our set-up, cells are sandwiched in a single plane between two collagen-fibronectin gels. This trick facilitates the analysis of cell tracks, which are for the most part 2D, at least in the beginning, but in a 3D environment. This protocol has been previously published in Visweshwaran et al. (2018) and is described here in more detail.
Keywords: Collagen gelBackground
The migration of cells within our body is an essential process and driven by complex underlying cellular functions. It plays an important role in many physiological processes, including development, immune responses, and tissue regeneration (Kunwar et al., 2006; Friedl and Weigelin, 2008; Reig et al., 2014). In addition, certain pathological situations such as tumor invasion and metastasis rely on cell motility (Thiery, 2002). Consequently, cell migration has become a major field of study in the context of both fundamental and translational research. Although in the last decade we have witnessed enormous advances in the understanding of the mechanisms underlying the highly plastic process of cell migration, many questions remain open. Especially, the complex regulation of migration is still unclear within differently composed microenvironments that are crowded by cells and extracellular matrix (Petrie et al., 2009; Friedl and Wolf, 2010).
To analyze cell migration, several in vitro and in vivo cell migration assays have been developed over the years. Thein vivo cell migration assays most closely mimic the real physiological situation and observe cells within their natural environment with its complexities of variable extracellular matrix (ECM) composition, geometry, topography and pore size. However, performing such experiments is labor- and cost-intensive, time-consuming, tough to control and requires advanced imaging techniques and animal experiments. Due to such practical challenges, cell migration has traditionally been studied on two-dimensional (2D) surfaces (Dang and Gautreau, 2018), e.g., in the context of wound-healing assays (Molinie and Gautreau, 2018). While this works to some extent for adherent cells such as breast epithelial carcinoma cells, 2D migration assays have little physiological relevance and thus little predictive value. Moreover, the striking difference between the 2D and the 3D settings becomes understandable in the light of recent studies of cell motility (Lämmermann et al., 2009; Friedl et al., 2012; Petrie and Yamada, 2016). These recent studies demonstrate that cell migration is a very plastic process and the cells embedded in 3D matrices composed of collagens or matrigel employ a very different locomotory machinery than cells on 2D surfaces. Therefore, studying the migration of cells embedded within a physiological-like 3D environment could lead to the results that have more significance and better predictive value.
Apart from being easier to perform than true in vivo migration experiments, 3D migration assays with their simpler matrix composition offer the advantage of a controlled, easily manipulable environment. Thus, it facilitates easy dissection of molecular mechanisms and the interpretation of experimental results. The 3D migration studies are usually done with the help of Boyden chambers (e.g., transwell assays [Visweshwaran et al., 2018]). However, these assays typically provide only an endpoint readout of cell migration efficiency, thereby not much information derived for an in-depth analysis. In contrast, real-time microscopy-based 3D assays allow one to observe and track individual cells, and thus the analysis of various parameters such as cell morphology during migration, cell speed, and directionality.
To our knowledge, there are not many optimal methods available for 3D cell migration analysis. All existing methods have their limitations. They either allow the experimenter to use a set-up where the embedded cells in the collagen gel are prone to migrate randomly in all directions including Z-axis, which render them hard to track and quantify migration parameters, or compel the experimenter to use complex, hard-to-handle and often costly setups (Rommerswinkel et al., 2014; Biswenger et al., 2018) to perform 3D migration assays. To overcome these limitations, we have developed a set-up where cells are sandwiched in a single plane between two collagen-fibronectin gels. This procedure facilitates the analysis of cell tracks, which are for the most part 2D, at least in the beginning, but in a 3D environment. In essence, here we describe an easy method for performing and analyzing 3D migration assays based on a home-made set-up and an easy analysis pipeline.
Materials and Reagents
- µ-slide 8 well with a glass bottom (Ibidi, catalog number: 80827)
- Cell line: MDA-MB-231 (ATCC, catalog number: HTB-26)
- L-15 medium (Thermo Fisher Scientific, catalog number: 11415049)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: A31605)
- Penicillin-Streptomycin (100x) (Thermo Fisher Scientific, catalog number: 15140122)
- Trypsin (0.25%) (Thermo Fisher Scientific, catalog number: 15050065)
- DPBS (Thermo Fisher Scientific, catalog number: 14190250)
- 10x MEM (Thermo Fisher Scientific, catalog number: 11430030)
- HEPES (Thermo Fisher Scientific, catalog number: 15630080)
- Collagen I (Rat-tail, 4.41 mg/ml) (BD Biosciences, catalog number: 354236)
- Fibronectin (5 mg) (FN) (Sigma, catalog number: F1141)
- MDA-MB-231 culture medium (see Recipes)
- Collagen-Fibronectin gel mix (see Recipes)
Equipment
- Phase contrast microscope with 20x air objective, temperature module, CO2 module and auto focus module (Zeiss Axio Observer microscope)
- 37 °C cell culture incubator
- Laminar airflow hood
Software
- Fiji: ImageJ with MTrackJ plugin
Note: Fiji: ImageJ, software could be downloaded from https://imagej.net/Fiji/Downloads and for MTrackJ plugin information go to https://imagescience.org/meijering/software/mtrackj/. - Microsoft (MS) Excel 2007 or later
Procedure
- Prepare the appropriate volume of collagen-fibronectin mix as described in Recipe 2.
- Add 100 µl of gel mix per well of µ-Slide 8 well and let it polymerize at 37 °C for 1 h (Figure 1A).
Note: After gelification, collagen-fibronectin gel becomes slightly milky white and collagen fibers become uniformly visible. - During this gelification period, trypsinize the cells and count them. Set the cell concentration to 104 cells per ml.
- After gel polymerizes, put 200 µl of medium containing cells (i.e., 2,000 cells) on top of the gel.
- Put µ-slide 8 well in the incubator at 37 °C and allow the cells to adhere on the gel surface for about 2 h (Figure 1A).
- Once cells have adhered, remove the medium gently. This can be done by slowly pipetting out the medium from the corners of the µ-slide well.
Note: This step is delicate. While removing the medium, extra care must be taken not to disturb the gel and cells. If the gel gets disturbed/damaged, then they are not good for imaging. - After removing the medium, keep the µ-slide aside in the laminar airflow hood and quickly, re-prepare the collagen-fibronectin mix.
- Put 100 µl of gel mix on the top of the cells adhering to the previously casted gel (Figure 1A).
- Incubate for 1 h at 37 °C in the incubator for the gel to polymerize.
Note: After gelification of the second layer of gel, one can observe cells that are sandwiched between two gels in a single z plane. - Meanwhile, switch on the microscope and the microscope stage heater. Adjust the temperature to 37 °C.
- After gelification of the second layer gel, add 150 µl of complete medium on top of the second gel to ensure the gels do not dry out (Figure 1A).
- Carefully place the µ-slide into a microscope insert designed to hold the slides and allowing for temperature, humidity, and CO2 control.
- Place the insert onto the microscope stage. Set the focus on the cells that are at the center of well. Use a 20x objective. Use an objective with phase contrast (Figure 1B).
Note: The center of the µ-slide wells is best for imaging to avoid aberrations close to the sides of the wells. - Acquire time-lapse images for 60 h with a time-lapse interval of 20 min.

Figure 1. Steps in 3D migration setup. A. Schematic overview depicting the steps involved in the protocol. B. Exemplary time-lapse images of the MDA-MB-231 cells that are sandwiched between two collagen-fibronectin gels, depicting the cell morphology changes during its 3D migration. Scale bar: 40 µm. For the exemplary depiction of the 3D migration video, refer to Visweshwaran et al., 2018, Movie EV2.
Data analysis
- Once the video acquisition is finished, assign the image properties. Open the video/image stack file in Fiji and assign the image properties by selecting ‘Image’ → ‘Properties…’ and fill in ‘Unit of length’ (example-µm), ‘Pixel width’ (pixel size in µm), ‘Pixel height’ (pixel size in µm) and ‘Frame interval’ (20 min). Save the video/image stack. A similar assignment can also be done by selecting ‘Analyze’ → ‘Set scale’ option.
- Manual tracking and analysis of cell trajectories are done as described in Dang and Gautreau, 2018, and Gorelik and Gautreau, 2014. Briefly, manual tracking performed with the help of Fiji software plugin 'MTrackJ', which gives the trajectory data. These data are then copied to MS Excel file containing DiPer suite of custom-made macros for quantifying migration parameters. By running the DiPer macros, various migration parameters like cell speed, directional persistence (direction autocorrelation), and mean squared displacement (MSD) are calculated and plotted, which are used to make publication-quality figures (Figure 2).
- Automated tracking and analysis of cell trajectories are also possible for this set-up. Visweshwaran and Maritzen, 2019 describes a suitable procedure to quantify 3D cell migration automatically with minimal input by the experimenter.
Note: MS Excel file loaded with DiPer suite of macros are available as supplementary file: Excel_loaded_with_DiPer. Detailed description about DiPer and the migration parameters like cell speed, directional persistence (direction autocorrelation) and mean squared displacement (MSD) available in Gorelik and Gautreau, 2014.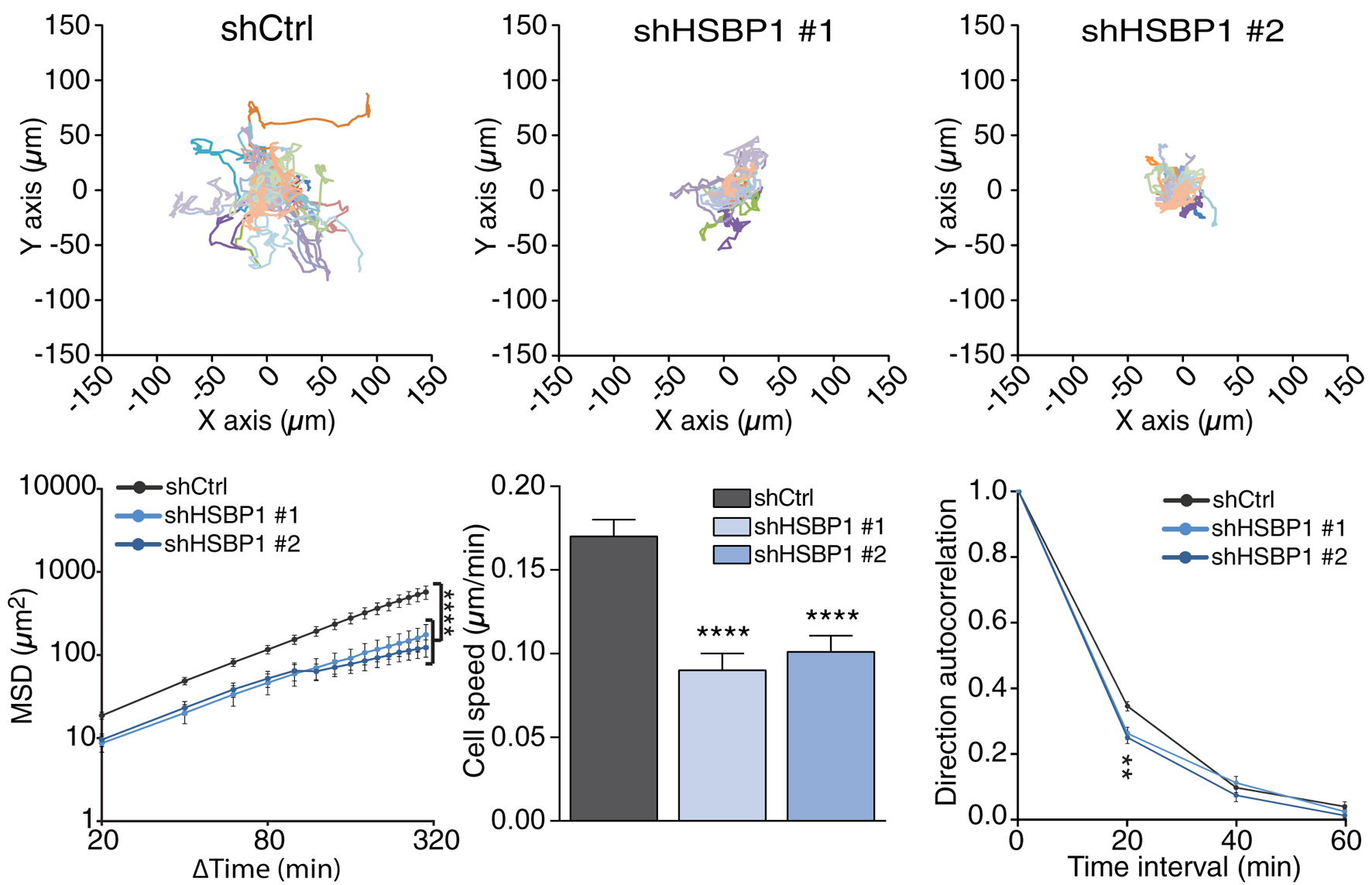
Figure 2. Representative 3D migration quantification of HSBP1 depleted MDA-MB-231 cells. Please refer to Visweshwaran et al. (2018) for details. Briefly, top panels display single-cell trajectories. Mean Square Displacement (MSD) analysis shows that HSBP1 depleted cells explore a smaller territory than control cells. This effect might be accounted for by decreased speed and directional persistence. At least 10 cells were tracked per condition, in 3 independent experiments; data are mean ± S.E.M; ANOVA **P < 0.01; ****P < 0.0001.
Notes
- When MDA-MB-231 cells are cultured in Leibovitz’s L-15 medium, no CO2 should be used for pH control. The L-15 medium formulation was devised for use with atmospheric air. Incubation in an atmosphere enriched with CO2 would be detrimental to cells.
- For running macros to analyze the cell trajectories, ‘Developer’ tab in the MS Excel must be enabled. The ‘Developer’ tab is enabled by going to ‘Excel Options’ → ‘Customize Ribbon’ → ‘Customize the Ribbon’ → ‘Main tab’ → checking ‘Developer’.
- The effects of drugs/inhibitors on 3D migration of the cells could be investigated in this set-up. For a long-term effect analysis, the drug could be added while making the collagen-fibronectin gels and in the medium that is added at the final stage to prevent the gel drying. In this method, cells are exposed constantly to the drug for a full period of observation. On the other end, the addition of drugs/inhibitors during 3D migration is also possible. In this method, a high concentration of drug should be added (1-10 µl) gently to the medium that is present on the top of the gels to prevent the gel drying. Here, the drug will diffuse into the gels. In this case, the experimenter initially would record a 3D cell migration for a period where there is no drug treatment and after addition of the drug, a period of 3D cell migration under drug treatment. If the drugs/inhibitors are toxic, a brief pre-treatment of the cells with the drug and sandwiching them between gels for migration analysis are also an alternative method.
Recipes
- MDA-MB-231 culture medium
L-15 medium
10% FBS
1x Penicillin-Streptomycin - Collagen-Fibronectin gel mix
2 mg/ml Collagen I
10 µg/ml FN
25 mM HEPES
10% FBS
1x MEM completed with sterile water
For 1 ml gel volume:10x MEM 100 µl HEPES (1 M) 25 µl Collagen I (4.41 mg/ml) 452 µl FBS 100 µl FN 10 µl Sterile water 313 µl
Acknowledgments
This protocol was adapted from Dang et al. (2013) with modifications. The establishment of this protocol was funded by AG’s group grant: the Agence Nationale de la Recherche (ANR ANR-15-CE13-0016-01), the fondation ARC pour la Recherche sur le Cancer (PGA120140200831), and Institut National du Cancer (INCA_6521). SPV was supported by PhD fellowships from Ministère de l'Enseignement Supérieur et de la Recherche for the first 3 years and from Ligue Nationale contre le Cancer for the 4th year.
Competing interests
The authors declare that they have no conflict of interests.
References
- Biswenger, V., Baumann, N., Jurschick, J., Hackl, M., Battle, C., Schwarz, J., Horn, E. and Zantl, R. (2018). Characterization of EGF-guided MDA-MB-231 cell chemotaxis in vitro using a physiological and highly sensitive assay system. PLoS One 13(9): e0203040.
- Dang, I. and Gautreau, A. (2018). Random migration assays of mammalian cells and quantitative analyses of single cell trajectories. Methods Mol Biol 1749: 1-9.
- Dang, I., Gorelik, R., Sousa-Blin, C., Derivery, E., Guerin, C., Linkner, J., Nemethova, M., Dumortier, J. G., Giger, F. A., Chipysheva, T. A., Ermilova, V. D., Vacher, S., Campanacci, V., Herrada, I., Planson, A. G., Fetics, S., Henriot, V., David, V., Oguievetskaia, K., Lakisic, G., Pierre, F., Steffen, A., Boyreau, A., Peyrieras, N., Rottner, K., Zinn-Justin, S., Cherfils, J., Bieche, I., Alexandrova, A. Y., David, N. B., Small, J. V., Faix, J., Blanchoin, L. and Gautreau, A. (2013). Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503(7475): 281-284.
- Friedl, P. and Weigelin, B. (2008). Interstitial leukocyte migration and immune function. Nat Immunol 9(9): 960-969.
- Friedl, P. and Wolf, K. (2010). Plasticity of cell migration: a multiscale tuning model. J Cell Biol 188(1): 11-19.
- Friedl, P., Sahai, E., Weiss, S. and Yamada, K. M. (2012). New dimensions in cell migration. Nat Rev Mol Cell Biol 13(11): 743-747.
- Gorelik, R. and Gautreau, A. (2014). Quantitative and unbiased analysis of directional persistence in cell migration. Nat Protoc 9(8): 1931-1943.
- Kunwar, P. S., Siekhaus, D. E. and Lehmann, R. (2006). In vivo migration: a germ cell perspective. Annu Rev Cell Dev Biol 22: 237-265.
- Lämmermann, T. and Sixt, M. (2009). Mechanical modes of 'amoeboid' cell migration. Curr Opin Cell Biol 21(5): 636-644.
- Molinie, N. and Gautreau, A. (2018). Directional collective migration in wound healing assays. Methods Mol Biol 1749: 11-19.
- Petrie, R. J. and Yamada, K. M. (2016). Multiple mechanisms of 3D migration: the origins of plasticity. Curr Opin Cell Biol 42: 7-12.
- Petrie, R. J., Doyle, A. D. and Yamada, K. M. (2009). Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol 10(8): 538-549.
- Reig, G., Pulgar, E. and Concha, M. L. (2014). Cell migration: from tissue culture to embryos. Development 141(10): 1999-2013.
- Rommerswinkel, N., Niggemann, B., Keil, S., Zanker, K. S. and Dittmar, T. (2014). Analysis of cell migration within a three-dimensional collagen matrix. J Vis Exp (92): e51963.
- Thiery, J. P. (2002). Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2(6): 442-454.
- Visweshwaran, S. P., Thomason, P. A., Guerois, R., Vacher, S., Denisov, E. V., Tashireva, L. A., Lomakina, M. E., Lazennec-Schurdevin, C., Lakisic, G., Lilla, S., Molinie, N., Henriot, V., Mechulam, Y., Alexandrova, A. Y., Cherdyntseva, N. V., Bieche, I., Schmitt, E., Insall, R. H. and Gautreau, A. (2018). The trimeric coiled-coil HSBP1 protein promotes WASH complex assembly at centrosomes. EMBO J 37(13): pii: e97706.
- Visweshwaran, S. P. and Maritzen, T. (2019). A simple 3D cellular chemotaxis assay and analysis workflow suitable for a wide range of migrating cells. MethodsX 6: 2807-2821.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Visweshwaran, S. P. and Gautreau, A. (2020). Analysis of Random Migration of Cancer Cells in 3D. Bio-protocol 10(1): e3482. DOI: 10.21769/BioProtoc.3482.
Category
Cancer Biology > Invasion & metastasis > Cell biology assays > Cell migration
Cancer Biology > Invasion & metastasis > Cell biology assays > Cell invasion
Cell Biology > Cell movement > Cell migration
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











