- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation and Characterization of Live Yeast Cells from Ancient Clay Vessels
(*contributed equally to this work) Published: Vol 10, Iss 1, Jan 5, 2020 DOI: 10.21769/BioProtoc.3473 Views: 6394
Reviewed by: David PaulFrancis FangMaría Victoria MartinAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Polyethylene Glycol-mediated Transformation of Drechmeria coniospora
Le D. He and Jonathan J. Ewbank
Mar 5, 2017 9451 Views

Isolation of Powdery Mildew Haustoria from Infected Barley
Linhan Li [...] Pietro D. Spanu
Jul 20, 2019 7831 Views

Development of an Efficient Transformation System for Halotolerant Yeast Debaryomyces hansenii CBS767
Anu P. Minhas and Dipanwita Biswas
Sep 5, 2019 4225 Views
Abstract
Ancient fermented food has been studied mainly based on residue analysis and recipes and reconstruction attempts were performed using modern domesticated yeast. Furthermore, microorganisms which participated in fermentation were studied using ancient-DNA techniques. In a recent paper, we presented a novel approach based on the hypothesis that enriched yeast populations in fermented beverages could have become the dominant species in storage vessels and their descendants could be isolated and studied today. Here we present a pipeline for isolation of yeast from clay vessels uncovered in archeological sites and transferred to the microbiology lab where they can be isolated and characterized. This method opens new avenues for experimental archeology and enables attempts to recreate ancient food and beverages using the original microorganisms.
Keywords: Ancient pottery vesselsBackground
Food ways and dietary habits, offer significant insight into subsistence strategies, connectivity and cultural and ideological choices of societies (Beaudry and Metheny, 2015). This direction of research is rather restricted when applied to ancient societies, as only residues of food have survived the absence of preservatives. Fragmentary evidence has impeded research of ancient fermented alcoholic beverages such as beer, wine, mead (honey wine) and kvass, which played a major role in daily life as well as in religious, social and political rituals of ancient societies (Joffe, 1998; Myers et al., 2000; Dietler, 2006; Legras et al., 2007; McGovern, 2009; Schiefenhövel and Macbeth, 2011; Hornsey, 2012; Parker and McCool, 2015).
Reconstruction of ancient fermented beverages has, until now, consisted of brewing modern ingredients according to ancient recipes or based on organic residue found in ancient containers or breweries. To achieve fermentation, previous efforts have used Saccharomyces cerevisiae, the domesticated commercial yeast species (McGovern, 2009; Stika, 2011). To the best of our knowledge, the study of ancient microorganisms, including bacteria (Donoghue et al., 2004; Andam et al., 2016), viruses (Muhlemann et al., 2018) and yeast (Cavalieri et al., 2003), was based on ancient DNA studies; no attempt has been made to isolate and reproduce microorganisms from containers used for the production of fermented beverages.
Hypothesizing that yeast can form stable colonies in clay nanopores, we were recently able to isolate live yeast cells from ancient containers thought to have contained beverages and oil, using the current protocol (Aouizerat et al., 2019). Following the isolation, we characterized the growth of the yeast, sequenced and analyze their full genome and used them to produce and analyze beer and mead (Aouizerat et al., 2019).
Thus, our research suggests that ancient microorganisms can be isolated from archaeological finds, facilitating the identification of additional ancient foods and beverages. It also promotes the field of “experimental archeology”: the recreation of the past, mainly of ancient fermented food (such as cheese and pickles) and beverages.
The following text presents a two-part protocol for the collection of sherds/vessels from archeological dig sites, as well as the isolation and characterization of microorganisms. Part I details guidelines for handling vessels intended for microorganisms’ isolation. Part II describes the isolation process. While this protocol was applied to yeast, it can be easily modified to apply to other microorganisms, such as bacteria.
Part I: Guidelines for the excavation and collection of clay vessels for microorganisms’ isolation (field work)
Materials and Reagents
- Lab gloves
- 50 ml Falcon tubes (MINIPLAST EIN SHEMER, catalog number: 835-050-21-111)
- Clean plastic container
- Aluminum foil
- Tube markers
Equipment
- Folding table
- Recording device/s including camera, tablet, laptop, and cellular phones
Procedure
Note: In sites where containers intended for isolation of microorganisms are expected, do not allow any kind of food or beverage in the excavation area (primarily fruit, vegetables, beer, yogurt, bread, pickles, cheese but also others).
- Before and while excavating
- Define vessels that are to be collected for microorganisms’ isolation. For example:
- Vessels that are clearly meant for storing, serving and drinking of beverages or food or, vessels carrying beverage-related specific features, such as strainers (Figure 1).
- Vessels suggested to have been previously used for the preparation of fermented beverages and food such as dairy products (e.g., churns).
- For more examples see also Aouizerat et al. (2019).
- Increase the awareness of the team so that they are prepared to collect vessels for microorganisms’ isolation based on the context of the site. For instance, in the case of the discovery of a drinking set in situ, or a possible production site.
- Make sure that no fermented food of any kind, including unfiltered beer, sourdough and wine are being consume in the site. In addition, it is recommended that workers wash their hands and faces before starting.

Figure 1. Putative beer jug. An example of a vessel thought to be a beer jug due to its shape and strainer.
- Define vessels that are to be collected for microorganisms’ isolation. For example:
- Excavation and sample handling in the field
- Identify vessels suitable for microorganisms’ isolation as soon as you can, in situ, before they are fully uncovered.
- Visually record the excavation process, and fully document the vessel (drawing, photography, textual description) within its context, prior to its extraction from the soil.
- Set up a folding table near the vessel and cover the table with clean aluminum foil.
- From now on, work on this vessel using disposable lab or cotton gloves.
- When the identified vessel is uncovered, decide if and how much of the vessel can be broken based on the policy of the excavators and government regulations.
- Immediately after the initial uncovering, take small pieces (5-6 cm) of the vessel and transfer them to 50 ml tubes using gloves at all times.
Note: Try to minimize vessel exposure time before the initial sampling. - Label the tubes with a tube marker and record the time, place, specific context, excavator, and type of sherd.
- Carefully, using laboratory gloves, excavate the vessels according to the excavation’s regular protocol.
Note: Only dedicated and guided personnel, preferably only one, should touch the sherds to avoid possible contamination. - While uncovering, identify parts of the vessel that are more likely to hold microorganisms such as the base, strainer and the side of the curve towards the neck. These parts of the vessel will be the focus of efforts from now on.
- Transfer the collected fragments to the table, keep them in plastic containers and label them accordingly.
Note: Do not wash the pieces meant for microorganisms’ isolation. - Label the other fragments of the uncovered vessel that are not to be sent to the lab with the same ID, to keep them for restoration and other analyses.
- Always keep some fragments unwashed in case the lab would need more fragments.
- Transfer the plastic containers with the sherds for microorganism analysis to the microbiology lab (see below).
Note: In specific cases, an entire vessel can be chosen to be taken to the lab. In that case, try to minimize the time of the vessel exposure and excavate it, record and transfer it to a plastic container as soon as possible.
Control vessels and control soil samples:
Note: Non-related controls are a key component in the analysis, to ensure that the isolated microorganisms originated in the vessel and are not an environmental contamination. - Using gloves, collect as many samples as possible of: sediments, stones, plants, and roots from the vessel’s surroundings. Store them in 50 ml plastic tubes.
- Identify vessels which in high probability are unrelated to microorganisms, such as burned cooking pots, small vessels/juglets (i.e., putative perfume containers), and other vessels, and process them as described above for the putative fermented food containers.
Note: Containers suspected to have held oily products should not be considered as controls, since yeast are major inhabitants of oil (Aouizerat et al., 2019).
- Delivery to the lab
- Using gloves at all times, transfer the tubes, containers and their documentation to the lab.
- Store in-doors at room temperature.
Part II: Isolation and characterization of yeast from environmental samples (in the lab)
Materials and Reagents
- For yeast isolation
- 50 ml sterile Falcon tubes (MINIPLAST EIN SHEMER, ref: 835-050-21-111)
- 15 ml sterile Falcon tubes (MINIPLAST EIN SHEMER, ref: 835-015-40-111)
- Lab strain of S. cerevisiae to serve as a control. We used Sefale S-04 (Fermentis)
- YPD broth (Difco, catalog number: bbl242820)
- Agar (BactoTM Agar, BD, catalog number: 214010)
- Selective agar plates: Sabouraux dextrose agar supplemented with chloramphenicol and gentamicin 4 mg/ml (NOVAmed Israel, catalog number: BA-114)
- For yeast DNA purification
- AMPure XP beads (New England BioLabs, catalog number: E6270)
- Acid washed glass beads (Sigma-Aldrich, catalog number: G4649)
- Ice cubes
- RNase A (10 mg/ml DNase- and Protease-free RNase, Thermo Fisher Scientific, catalog number: EN0531)
- Sterile ddH2O
- Phenol:chlorophorm:Isoamyl alcohol in ratios of is 25:24:1 respectively, saturated with Tris (10 mM) and EDTA (1 mM) at pH 8 (Sigma-Aldrich, catalog number: P3803)
- Tris-HCl, pH 8.0 (10 mM), EDTA, pH 8.0 (TE buffer, Thermo Fisher Scientific, catalog number: AM9858)
- Ammonium acetate (Sigma-Aldrich, catalog number: A1542)
- Ethanol (GADOT, Israel)
- Nextera XT DNA kit (Illumina, catalog number: FC-131-1096)
- Malt extract (Chen shmuel chemicals, Israel)
- Hops (Hopsteiner, Hop pellets 90)
- Glycerol (50%) (Bio-Lab)
- PBS buffer
- “Smash and Grab” lysis solution (Harju et al., 2004) (see Recipes)
Equipment
- Lab Glass flasks
- Pipettes of all sizes (Eppendorf)
- Incubator that can heat to 30 °C (QSR Technologies)
- Incubator Shaker (New Branswick scientific, Exella E24 incubator shaker)
- Spectrophotometer (VWR, v-1200 spectrophotometer)
- Centrifuge (Eppendorf, centrifuge 5430R)
- Plate reader–SynergyTM 2 Multi-Mode Microplate Reader (BioTek)
- DNA quantification device such as Qubit (Invitrogen, Qubit® 2.0 fluorometer)
- -80 °C freezer (Thermo ScientificTM, catalog number: 5IDTSX)
- Vortex (Fisher Scientific, Fisher STD mini vortexer 230v)
- Fume hood of any kind for the work with the Phenol:chlorophorm:Isoamyl alcohol
- Laminar hood (Thermo ScientificTM, MSC-AdvantageTM)
- PCR hood (Lab Companion)
- PCR thermocycler (QSR Technologies, Thermal cycler GE48529)
- Microscope (ZEISS, AXIO Lab.A1)
- Microscope camera attached to PC (ZEISS, AXIOCAM 105 color)
Software
- 96-well plate reader software (BioTek, Gen5 2.07)
- GraphPad Prism 8.0.2 (https://www.graphpad.com/)
- Geneious Pro 2019 (https://www.geneious.com/)
- Microscope software (ZEISS, ZEN2 blue edition)
- Trimmomatic 0.36 (Bolger et al., 2014)
- FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc)
- Celera assembler 8.3rc2 (Myers et al., 2000)
- BlobTools V1 (Laetsch and Blaxter, 2017)
- R (https://www.r-project.org) with "growthcurver” package (https://cran.r-project.org/web/packages/growthcurver/vignettes/Growthcurver-vignette.html)
Procedure
Note: Work routinely with gloves and lab safety equipment (i.e., lab-coat, protective eye goggles). The beginning steps do not need sterile work conditions. However, after the initial isolation of the yeast, work in a laminar hood or near a burner.
- Yeast isolation from vessels and control samples
- Put the sherds in a plastic container and cover them with sterile liquid YPD medium.
- Incubate at room temperature without shaking for 7 days.
- Spread 0.1 ml samples from the medium on selective agar plates for fungal isolation and incubate at 30 °C for 12 to 48 h.
- Inspect the plates frequently for the appearance of yeast colonies (Figure 2A).
Note: Yeast colonies usually grow faster than molds and filamentous fungi. Do not wait for more than 48 h or the plate will be covered by molds and filamentous fungi.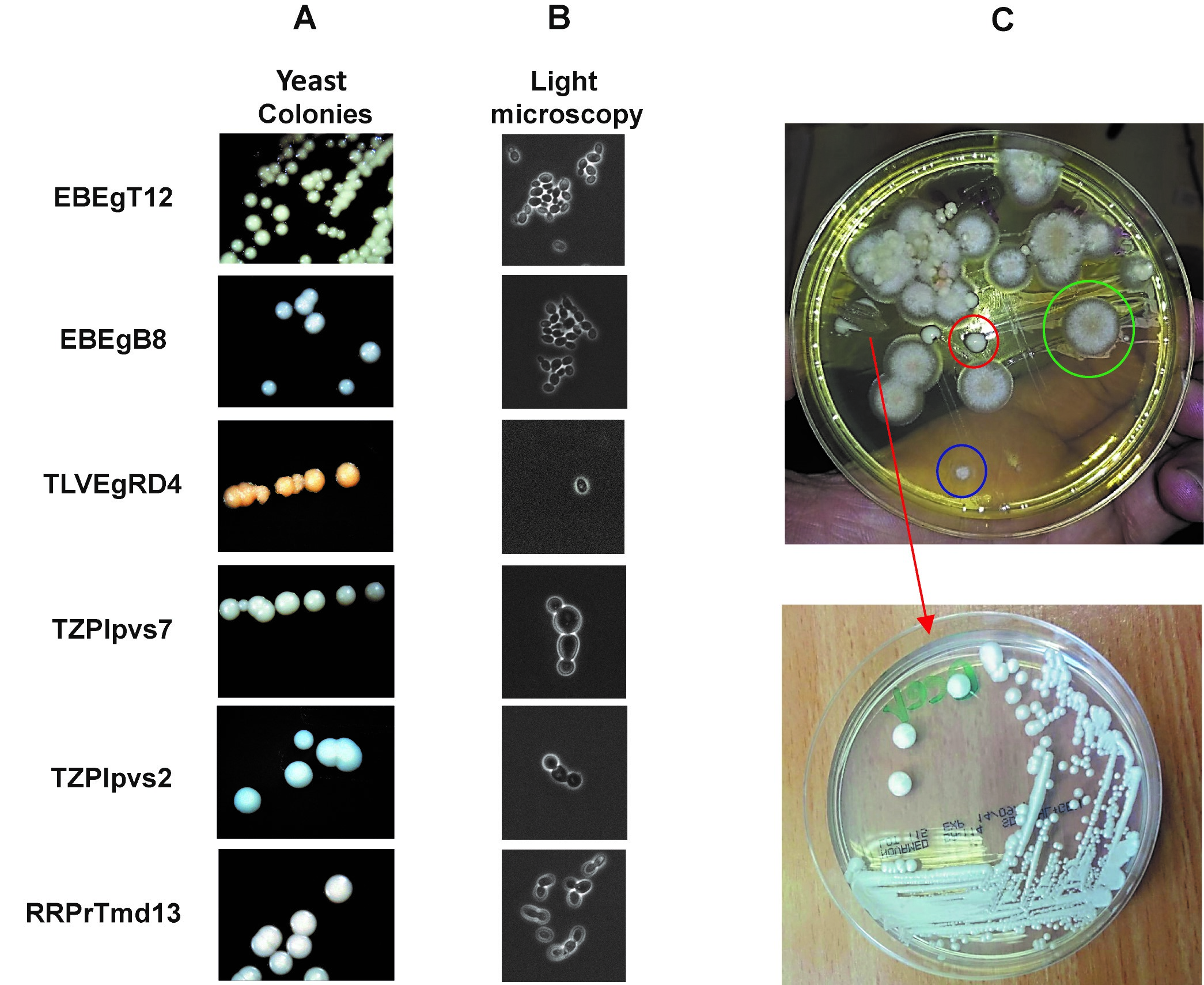
Figure 2. Yeast colonies and cells. Yeast have typical colonies and cell morphology both of which are distinct from molds and bacteria. A. several types of yeast colonies isolated from ancient vessels (Aouizerat et al., 2019). In general, yeast colonies are relatively large with a shiny white color in contrast to bacterial colonies which are usually small and matte. B. Yeast cells observed under light microscopy (60x). Note that yeast cells are usually bigger than typical bacteria and many kinds of yeast divide by budding rather than fission. C. Upper panel, a selective agar plate for fungal isolation was plated as described in the Procedure A of Part II. Note the low number of bacterial colonies due to the antibiotics (blue circle), structure of molds (green circle) and distinctive shiny colony of yeast (red circle). In the bottom panel, a yeast colony which was picked and re-streaked on a YPD plate (red arrow). - Collect any colony which seems to be a yeast colony, observe for typical yeast cells using light microscopy (Figure 2B) and plate it on solid YPD agar plates, containing 2% w/v Bacto-agar (Figure 2C).
- Incubate the plates at 30 °C for 24-48 h until yeast colonies are clearly observed.
Note: If a mixture of molds and yeast grow, re-pick a separate yeast colony to a new YPD plate and repeat Step A6. - Inspect the colonies visually and under light microscope (60x magnification) for typical yeast cells which are distinct from bacteria colonies (Figure 2).
- To make a frozen stock of the yeast, pick a single colony to 5 ml YPD broth and incubate it for 24 h at 30 °C, in a shaker incubator under aerobic conditions with agitation (250-300 rpm).
- Mix 0.5 ml of the culture with sterile 0.5 ml glycerol (50%), vortex to mix, aliquot and store at -80 °C.
- Yeast Characterization
- Yeast DNA isolation
Note: Yeast cell DNA isolation was performed as previously described (Holm et al., 1986).- Select a yeast containing tube from the -80 °C and allow to defrost at room temperature. When thawed, perform isolation streak on a YPD plate and incubate at 30 °C until colonies are observed (about 48 h).
- Transfer a single colony to 10 ml YPD broth in a sterile 100 ml flask. Grow the yeast overnight at 30 °C with 200 rpm shaking.
- Transfer the culture to 50 ml Falcon tube and centrifuge at 1,000 x g for 5 min. Pour out the supernatant, re-suspend the pellet in sterile water, re-centrifuge and discard the supernatant.
- In a fume hood, add 200 µl of Phenol:chlorophorm:Isoamyl alcohol, 0.3 g acid washed glass beads and 200 µl of Smash and Grab solution and vortex for 3 min in order to lyse the cells.
- Add 0.2 ml TE buffer and centrifuge the cells at 1,000 x g for 5 min at room temperature.
- Transfer the aqueous layer containing the DNA to 1 ml ethanol and incubate it for 20 min at -80 °C.
- Wash by centrifugation at 10,000 x g for 5 min and re-suspend in 0.4 ml TE buffer.
Note: DNA pellet is transparent and can be difficult to see–avoid losing the pellet. - Add 1 µl of RNase A.
- Incubate the solution at 37 °C for 5 min.
- Add 10 µl of ammonium acetate (4 M) and 1ml of 70% ethanol and incubate for 20 min at -80 °C.
- Centrifuge at maximal speed for 10 min (at least 15K x g) and discard the supernatant.
- Re-suspend the pellet in 50 µl of TE buffer.
- Quantify the DNA using Qubit, it should be 10-20 µg/ml.
- Store the extracted DNA at -20 °C.
- Yeast identification by internal transcribed spacer (ITS)
Note: For rapid and cheap identification you can use sanger sequencing of the ITS region. Keep in mind that the specific yeast species might not be represented in the database and thus full genome sequencing is preferable.- Amplify the ITS region using Standard PCR kit and program with Illumina’s primers: ITS1F; ACACTGACGACATGGTTCTACACTTGGTCATTTAGAGGAAGTAA and ITS2R; TACGGTAGCAGAGACTTGGTCTGCTGCGTTCTTCATCGATGC.
- Send the PCR fragments for Sanger sequencing.
- BLAST the sequence results against the ISHAM barcoding database (http://its.mycologylab.org) and the NCBI database (https://blast.ncbi.nlm.nih.gov/ Blast.cgi).
- Full genome sequencing and assembly
Note: DNA sequencing can be performed using any common NGS technology. Our libraries were prepared using a Nextera XT DNA kit (Illumina, San Diego, CA), and the DNA was amplified by a limited-cycle PCR followed by purification using AMPure XP beads. The DNA libraries were normalized, pooled, and tagged in a common flow cell at 2 x 250 base-paired-end reads using the NextSeq platform. For more detailed protocols see Aouizerat et al. (2019). The assembly also can be performed using numerous NGS analysis tools. Here we describe the analysis we performed (Aouizerat et al., 2019).- Remove Illumina adaptors using Trimmomatic 0.36.
- Determine the quality of the reads using FastQC.
- Perform De-novo assembly with the Celera assembler 8.3rc2.
- Exclude non-target-species scaffolds using BlobTools V1.
- Growth curve analysis (in wort)
Note: This procedure is used for the comparison of various growth parameters of yeast or bacteria in liquid under various conditions such as varying media, temperatures and addition of different compounds. Here, the growth of yeast in beer wort was performed, in order to test whether the isolated yeast might have been selected for in the past under such conditions. As a positive control we used a modern beer-producing strain of S. cerevisiae which is commercially available and as a negative control the pathogen yeast Candida parapsilosois which does not produce beer.
Wort Preparation
Note: The wort is prepared according to a standard recipe (Kunze and Manger, 2004).- Heat water (5 L) to a pasteurization temperature of 72 °C.
- Add malt extract to a final concentration of 100 g/L, while thoroughly stirring, and allow to infuse together for 30 min in temperatures between 63 and 67 °C.
- Increase the solution temperature to 100 °C.
- Add 1 g/L of hops.
- Boil the solution for 45 min.
- Meanwhile prepare 5 L ice-cold water by mixing water with ice cubes.
- Add 1 g/L of hops once more.
- Let boil for an additional minute.
- Add the previously prepared ice-cold water (from Section f) to the mixture.
- Transfer the wort to a sterile flask and keep at 4 °C.
- Perform isolation streak of yeast from frozen stock on YPD agar plate.
- Incubate the plate at 30 °C for 48 h.
- Transfer a colony to 20 ml liquid YPD and incubate it at 30 °C, under aerobic conditions with agitation (250-300 rpm).
- Centrifuge the yeast at 1,000 x g for 5 min, wash in sterile PBS buffer and re-suspend the pellet in wort.
- Dilute the solution 1:100 in wort.
- Add 200 μl of the diluted solution/well of 96-well plate in triplicates or more repeats.
- Incubate the plate in a plate reader at 30 °C with linear shaking of 5 s every 20 min followed by an optical density (OD600) reading for 48-96 h.
- Export the results to a text file (Figure 3A).
- Fit a logistic equation using R package “growthcurver” (Figure 3B).
- Compare the statistical significance of the differences between the various parameters using Student's t-test.
- Determine the relative distance between the samples using Principal Components Analysis (PCA, R prcomp () command) (Figure 3C).
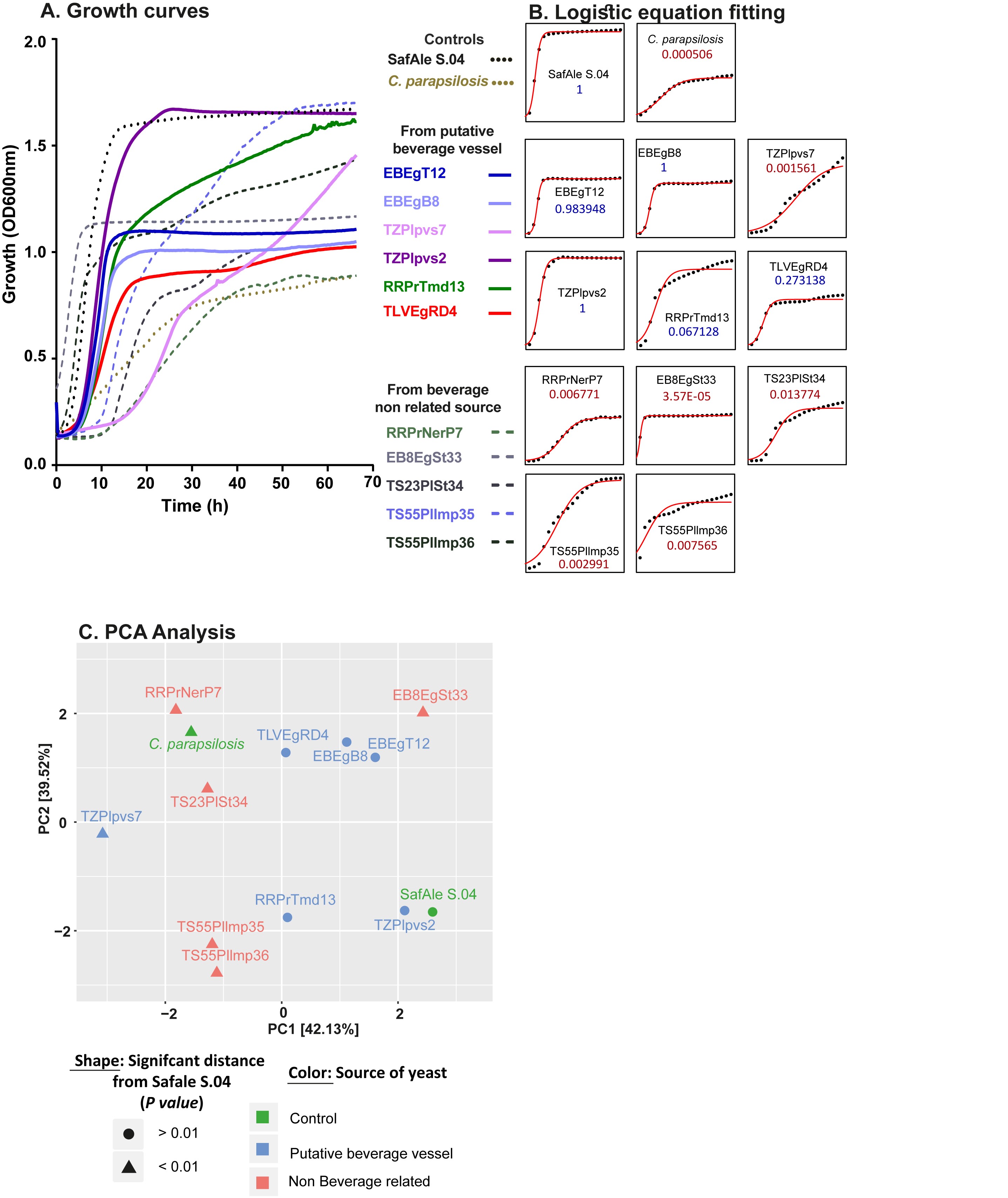
Figure 3. Example of growth curve analysis. A. Growth curves were read as output from the plate reader. B. A logistic curve was fitted to each curve using R growthcurver. C. A PCA representation of the distance between the various parameters of the logistic equations of each sample. These results were adapted from Aouizerat et al., 2019.
- Yeast DNA isolation
Notes
For DNA sequencing we enlisted the help of the interdepartmental equipment unit of the Ein-Kerem Campus, Hebrew University of Jerusalem.
Recipes
- “Smash and Grab” lysis solution
Triton X-100 (2%)
SDS (1%)
NaCl (100 mM)
Tris-HCl, pH 8.0 (10 mM)
EDTA, pH 8.0 (1 mM)
Acknowledgments
This protocol was adapted from Aouizerat et al. (2019). Isolation and characterization of live yeast cells from ancient vessels as a tool in bio-archeology. mBio 10(2): e00388-19. We thank Prof. Christina Warinner from University of Jena for her helpful suggestions.
Competing interests
Conflict of interest statement: None declared.
References
- Andam, C. P., Worby, C. J., Chang, Q. and Campana, M. G. (2016). Microbial Genomics of Ancient Plagues and Outbreaks. Trends Microbiol 24(12): 978-990.
- Aouizerat, T., Gutman, I., Paz, Y., Maeir, A. M., Gadot, Y., Gelman, D., Szitenberg, A., Drori, E., Pinkus, A., Schoemann, M., Kaplan, R., Ben-Gedalya, T., Coppenhagen- Glazer, S., Reich, E., Saragovi, A. S., Lipschits, O., Klutstein, M. and Hazan, R. (2019). Isolation and characterization of live yeast cells from ancient vessels as a tool in bio-archaeology. mBio 10(2): e00388-19.
- Beaudry, M. C. and Metheny, K. B.(2015). Archaeology of food: an encyclopedia. Rowman & Littlefield.
- Bolger, A. M., Lohse, M. and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15): 2114-2120.
- Cavalieri, D., McGovern, P. E., Hartl, D. L., Mortimer, R. and Polsinelli, M. (2003). Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol 57 Suppl 1: S226-232.
- Dietler, M. (2006). Alcohol: anthropological/archaeological perspectives. Annu Rev Anthropol 35: 229-249.
- Donoghue, H. D., Spigelman, M., Greenblatt, C. L., Lev-Maor, G., Bar-Gal, G. K., Matheson, C., Vernon, K., Nerlich, A. G. and Zink, A. R. (2004). Tuberculosis: from prehistory to Robert Koch, as revealed by ancient DNA. Lancet Infect Dis 4(9): 584-592.
- Harju, S., Fedosyuk, H. and Peterson, K. R. (2004). Rapid isolation of yeast genomic DNA: Bust n' Grab. BMC Biotechnol 4: 8.
- Hornsey, I. S. (2012). Alcohol and its role in the evolution of human society. Royal Society of Chemistry.
- Holm, C., Meeks-Wagner, D. W., Fangman, W. L. and Botstein, D. (1986). A rapid, efficient method for isolating DNA from yeast. Gene 42(2): 169-173.
- Joffe, A. H. (1998). Alcohol and social complexity in ancient western Asia. Curr Anthropol 39: 297-322.
- Kunze, W. and Manger, H. J. (2004). Technology brewing and malting. VLB, Berlin, Germany.
- Laetsch, D. R. and Blaxter, M. L. (2017). BlobTools: interrogation of genome assemblies. F1000 Res 6: 1287.
- Legras, J. L., Merdinoglu, D., Cornuet, J. M. and Karst, F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol 16(10): 2091-2102.
- McGovern, P. E. (2009). Uncorking the past: the quest for wine, beer, and other alcoholic beverages. University of California Press.
- Muhlemann, B., Jones, T. C., de Barros Damgaard, P., Allentoft, M. E., Shevnina, I., Logvin, A., Usmanova, E., Panyushkina, I. P., Boldgiv, B., Bazartseren, T., Tashbaeva, K., Merz, V., Lau, N., Smrcka, V., Voyakin, D., Kitov, E., Epimakhov, A., Pokutta, D., Vicze, M., Price, T. D., Moiseyev, V., Hansen, A. J., Orlando, L., Rasmussen, S., Sikora, M., Vinner, L., Osterhaus, A., Smith, D. J., Glebe, D., Fouchier, R. A. M., Drosten, C., Sjogren, K. G., Kristiansen, K. and Willerslev, E. (2018). Author Correction: Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature 562(7726): E4.
- Myers, E. W., Sutton, G. G., Delcher, A. L., Dew, I. M., Fasulo, D. P., Flanigan, M. J., Kravitz, S. A., Mobarry, C. M., Reinert, K. H., Remington, K. A., Anson, E. L., Bolanos, R. A., Chou, H. H., Jordan, C. M., Halpern, A. L., Lonardi, S., Beasley, E. M., Brandon, R. C., Chen, L., Dunn, P. J., Lai, Z., Liang, Y., Nusskern, D. R., Zhan, M., Zhang, Q., Zheng, X., Rubin, G. M., Adams, M. D. and Venter, J. C. (2000). A whole-genome assembly of Drosophila. Science 287(5461): 2196-2204.
- Parker, B. J. and McCool, W. (2015). Indices of household maize beer production in the Andes: an ethnoarchaeological investigation. J Anthropol Res 71(3): 359-400.
- Schiefenhövel, W. and Macbeth, H. (2011). Liquid bread: beer and brewing in cross-cultural perspective. Berghahn Books.
- Stika, H. P. (2011). Early Iron age and late mediaeval malt finds from Germany—attempts at reconstruction of early Celtic brewing and the taste of Celtic beer. Archaeol Anthrop Sci 3(1): 41-48.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Aouizerat, T., Maeir, A. M., Paz, Y., Gadot, Y., Szitenberg, A., Alkalay-Oren, S., Coppenhagen-Glazer, S., Klutstein, M. and Hazan, R. (2020). Isolation and Characterization of Live Yeast Cells from Ancient Clay Vessels. Bio-protocol 10(1): e3473. DOI: 10.21769/BioProtoc.3473.
Category
Microbiology > Microbial genetics > Transformation
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










