- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Differentiation of Human iPSC-derived Retinal Pigment Epithelium Cells (iPSC-RPE)
Published: Vol 9, Iss 24, Dec 20, 2019 DOI: 10.21769/BioProtoc.3469 Views: 8034
Reviewed by: Giusy TornilloTomohiro MizutaniAgnieszka Maria Pastula

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2432 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 272 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 211 Views
Abstract
Induced Pluripotent Stem Cells (iPSCs) serve as an excellent model system for studying the molecular underpinnings of tissue development. Human iPSC-derived retinal pigment epithelium (iPSC-RPE) cells have fetal-like molecular profiles. Hence, biobanks like iPSCORE, which contain iPSCs generated from hundreds of individuals, are an invaluable resource for examining how common genetic variants exert their effects during RPE development resulting in individuals having different propensities to develop Age-related Macular Degeneration (AMD) as adults. Here, we present an optimized, cost-effective and highly reproducible protocol for derivation of human iPSC-RPE cells using small molecules under serum-free condition and for their quality control using flow cytometry and immunofluorescence. While most previous protocols have required laborious manual selection to enrich for iPSC-RPE cells, our protocol uses whole culture passaging and yields a large number of iPSC-RPE cells with high purity (88-98.1% ZO-1 and MiTF double positive cells). The simplicity and robustness of this protocol would enable its adaption for high-throughput applications involving the generation of iPSC-RPE samples from hundreds of individuals.
Keywords: Human induced pluripotent stem (hiPSC)Background
Age-related macular degeneration (AMD) is a leading cause of vision loss in developed countries affecting 11 million individuals in the United States and about 170 million worldwide (Pennington and DeAngelis, 2016). Moreover, considering age as a main factor, in our current aging society, the incidence of AMD is estimated to increase to 198 million in 2020 and 288 million by 2040 and to 22 million in the United States alone by 2050 (Wong et al., 2014, Pennington and DeAngelis, 2016). Current therapeutic strategies, although effective, are expensive and limited to delaying the speed of disease progression, and AMD still eventually leads to a complete loss of vision (Al-Zamil and Yassin, 2017; Mitchell et al., 2018). At the time of preparation of this article, there are seven active clinical trials aimed to evaluate the effectiveness and optimize the conditions of the transplantation of the human iPSC-RPEs or human embryonic stem cell-derived retinal pigment epithelium (ESC-RPE) cells (NIH-ClinicalTrials.gov). Thus, the development of a robust and cost-effective method for generating large amounts of high quality iPSC-RPEs is imperative for the advancement of future therapeutic treatments of AMD and potentially other eye diseases in humans as well as domestic animals (Sparrow et al., 2010).
We have previously demonstrated the utility of employing iPSC-RPE cells to identify and study genetic variants playing a role in the development of AMD (Smith et al., 2019). In this study, we derived iPSC-RPE cells from six individuals (3 European Americans, 2 East Asian Americans, and 1 African American) and showed that they have morphological and molecular characteristics similar to those of naïve RPE cells. We showed that iPSC-RPE gene expression profiles are highly similar to that of human fetal RPE, and that their ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) peaks are enriched for relevant transcription factor motifs. We performed fine mapping of AMD risk loci integrating the molecular data from iPSC-RPE cells that resulted in the prioritization of four variants, including a potential regulatory SNP (rs943080) near VEGFA and a coding SNP (rs34882957, pP167S) in the C9 gene (Smith et al., 2019). Our findings illustrate that iPSC-RPE cells are an excellent model system to study the molecular functions of genetic variation associated with AMD.
The initial protocols for deriving iPSC-RPE cells developed over a decade ago involved spontaneous differentiation and were highly inefficient requiring the manual separation of pigmented iPSC-RPE patches from non-differentiated cells (Klimanskaya et al., 2004; Vugle et al., 2008). Subsequently, induced directed differentiation protocols resulted in higher yielded and quality of iPSC-RPE cells but required numerous animal-derived components and thus had relatively low reproducibility or were very time consuming (Aoki et al., 2006; Osakada et al., 2008; Buchholz et al., 2009; Idelson et al., 2009; Reichman et al., 2014; Hazim et al., 2017). The introduction of small molecules into iPSC-RPE differentiation protocols greatly simplified the procedure and resulted in higher reproducibility (Osakada et al., 2009; Maruotti et al., 2013; Maruotti et al., 2015). Previous studies have optimized differentiation protocols to derive iPSC-RPEs from a limited number of iPSC or ESC lines and in most cases utilized small format culture vessels. Here, we present an optimized protocol for deriving iPSC-RPEs from multiple human iPSC lines in large sized culture flasks. It is similar to the protocol of Maruotti et al. (2015), but initiates differentiation when iPSCs reach 80% confluency, which we found to be optimal for all tested iPSCORE lines (Panopoulos et al., 2017). Additionally, we modified the length of time of exposure to small molecules at two steps, which resulted in further increase of yield and purity of iPSC-RPEs (88-98.1% ZO-1 and MiTF double positive cells). Our optimized protocol allowed us to derive iPSC-RPEs from six iPSC lines generated from ethnically diverse individuals under identical culturing conditions without the requirement of any individualized optimization steps.
Materials and Reagents
- iPSC Cell Culture
- 6-well plates (Corning, catalog number: 3506)
- Syringe filter 0.2 μm (VWR, catalog number: 28145-501)
- Soft-Ject® 3-Part Disposable Syringe, Air-Tite-3 ml (VWR, catalog number: 89215-234)
- 5 ml Borosilicate serological pipettes (Fisher Scientific, catalog number: 1367827E)
- 5 ml Serological pipettes (Bio Pioneer, catalog number: GEX0050-S01)
- 10 ml Serological pipettes (Bio Pioneer, catalog number: GEX0100-S01)
- P20 pipette tips sterile with filter
- P1000 pipette tips sterile with filter
- 15 ml conical tubes (Bio Pioneer, catalog number: CNT-15R)
- iPSC cells
- 70% ethanol
- UltraPureTM DNase/RNase-Free Distilled Water (Thermo Fisher Scientific, catalog number: 10977023)
- Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix (Corning, catalog number: 354230)
- mTeSRTM1 (Stem Cell Technologies, catalog number: 85850)
- DMEM/F-12 medium (Thermo Fisher Scientific, catalog number: 11330057)
- Dispase II (Thermo Fisher Scientific, catalog number: 17105041)
- Matrigel solution (Matrigel) (see Recipes: Table 1)
- 10 mM ROCK inhibitor, Y-27632 dihydrochloride solution (ROCK Inhibitor) (see Recipes: Table 2)
- 10x Dispase (see Recipes: Table 3)
- mTeSRTM1 complete medium (mTeSR) (see Recipes: Table 4)
- Monolayer plating
- 100 mm tissue culture dishes (Corning, catalog number: 430167)
- Automated cell counter slides (Bio-Rad Laboratories, catalog number: 1450019) or a hemocytometer (Hausser Scientific, catalog number: 1483) or equivalent
- 5 ml Serological pipettes (Bio Pioneer, catalog number: GEX0050-S01)
- 10 ml Serological pipettes (Bio Pioneer, catalog number: GEX0100-S01
- P20 pipette tips sterile with filter
- P200 pipette tips sterile with filter
- P1000 pipette tips sterile with filter
- 15 ml conical tubes (Bio Pioneer, catalog number: CNT-15R)
- 50 ml conical tubes (Bio Pioneer catalog number: CNT-50R)
- 70% ethanol
- Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix (Matrigel) (Corning, catalog number: 354230)
- mTeSRTM1 (Stem Cell Technologies, catalog number: 85850)
- DMEM/F-12 medium (Thermo Fisher Scientific, catalog number: 11330-057)
- Accutase (Innovative Cell Technologies, Inc., catalog number: AT 104)
- Trypan Blue Solution, 0.4% (Thermo Fisher Scientific, catalog number: 15250061)
- ROCK inhibitor, Y-27632 dihydrochloride (Selleck hem, catalog number: S1049)
- iPSC cell culture
- Matrigel solution (see Recipes: Table 1)
- 10 mM ROCK inhibitor, Y-27632 dihydrochloride solution (see Recipes: Table 2)
- mTeSRTM1 complete medium (see Recipes: Table 4)
- iPSC-RPE differentiation
- 100 mm tissue culture dishes (Corning, catalog number: 430167)
- (Optional) T150 tissue culture flasks, vented (Sigma, catalog number: Z707929)
Note: At the time of preparation of this manuscript Z707929 was no longer available. The same flasks are available under the catalog number Z707511-36EA (Sigma, catalog number Z707511-36EA). - 70 μm strainers (Fisher Scientific, catalog number: 431751)
- Automated cell counter slides (Bio-Rad Laboratories, catalog number: 1450019) or a hemocytometer (Hausser Scientific, catalog number: 1483) or equivalent
- 10 ml Serological pipettes (Bio Pioneer, catalog number: GEX0100-S01)
- 25 ml Serological pipettes (Bio Pioneer catalog number: GEX250-S01)
- 50 ml Serological pipettes (Bio Pioneer, catalog number: GEX500-S01)
- P20 pipette tips sterile with filter
- P200 pipette tips sterile with filter
- P1000 pipette tips sterile with filter
- Cell scraper (VWR International, catalog number: 179707)
- 15 ml conical tubes (Bio Pioneer, catalog number: CNT-15R)
- 50 ml conical tubes (Bio Pioneer, catalog number: CNT-50R)
- Nalgene Cryogenic vials (Thermo Fisher Scientific, catalog number: 5000-1020)
- iPSCs monolayer
- 70% ethanol
- Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix (Matrigel) (Corning, catalog number: 354230)
- DMEM/F-12 medium (Thermo Fisher Scientific, catalog number: 11330057)
- DMEM medium (Thermo Fisher Scientific, catalog number: 11965092)
- Ham’s F12 Nutrient Mix (Thermo Fisher Scientific, catalog number: 11765054)
- 1x Dulbecco’s phosphate buffered saline (DPBS) without calcium and magnesium (Thermo Fisher Scientific, catalog number: 14190250)
- B27 Supplement (50x), serum free (Thermo Fisher Scientific, catalog number: 17504044)
- KnockOutTM Serum Replacement (KOSR) (Thermo Fisher Scientific, catalog number: 10828028)
- L-Glutamine 200 mM (Thermo Fisher Scientific, catalog number: 25030081)
- MEM Non-Essential Amino Acids Solution 100x (Thermo Fisher Scientific, catalog number: 11140050)
- Penicillin-Streptomycin (10,000 U/ml) (Thermo Fisher Scientific, catalog number: 15140122)
- β-Mercaptoethanol (Thermo Fisher Scientific, catalog number: 21985023)
- Accutase (Innovative Cell Technologies, Inc., catalog number: AT 104)
- Nicotinamide (Sigma, catalog number: N3376)
- Chetomin (Sigma, catalog number: C9623)
- Trypan Blue Solution, 0.4% (Thermo Fisher Scientific, catalog number: 15250061)
- Dimethyl Sulfoxide (DMSO) (Sigma, catalog number: D2650-100ML)
- Liquid nitrogen
- RPE DM medium (see Recipes: Table 5)
- RPE medium (see Recipes: Table 6)
- 2x iPSC-RPE freezing medium (see Recipes: Table 7)
- 1 mM Chetomin solution (see Recipes: Table 8)
- 1 M Nicotinamide (100x) solution (see Recipes: Table 9)
- Flow cytometry
- 96-well round bottom assay plates (Genesee Scientific, catalog number: 25-224)
- CorningTM FalconTM Test Tube with Cell Strainer Snap Cap (Fisher Scientific, catalog number: 352235)
- CorningTM CostarTM Sterile Disposable Reagent Reservoirs (Fisher Scientific, catalog number: 4870)
- 5 ml Serological pipettes (Bio Pioneer, catalog number: GEX0050-S01)
- 10 ml Serological pipettes (Bio Pioneer, catalog number: GEX0100-S01)
- P20 pipette tips sterile with filter
- P200 pipette tips sterile with filter
- P1000 pipette tips sterile with filter
- P20 pipette tips without filter
- (Optional) P200 pipette tips without filter
- Fixation/Permeabilization Solution Kit with BD GolgiStopTM (BD Biosciences, catalog number: 554715)
- 1x Dulbecco’s phosphate buffered saline (DPBS) without calcium and magnesium (Thermo Fisher Scientific, catalog number: 14190250)
- Bovine Serum Albumin (BSA) (Sigma, catalog number: A2153-100G)
- (Optional) NaN3 (Sigma, catalog number: S2002-5G)
- 37% Formaldehyde (Sigma, catalog number: F-1635-500ML)
- Rabbit polyclonal anti-ZO-1 antibody (abcam, catalog number: ab59720)
Note: During preparation of this manuscript the antibody ab59720 was no longer available. A potential replacement: abcam, catalog number: ab221547. - Mouse monoclonal anti-MiTF antibody (abcam, catalog number: ab12039)
- Recombinant Rabbit IgG, monoclonal [EPR25A]–Isotype Control (abcam, catalog number: ab172730)
- Mouse IgG1, kappa monoclonal [15-6E10A7]–Isotype Control (abcam, catalog number: ab170190)
- Donkey-anti-Rabbit Alexa FluorTM 647 conjugated antibody (abcam, catalog number: ab150075)
- Goat-anti-Mouse Alexa FluorTM 488 conjugated antibody (Thermo Scientific, catalog number: A-11001)
- FACS Buffer (see Recipes: Table 10)
- FACS-FIX Buffer (see Recipes: Table 11)
- Immunofluorescence
- Millicell EZ SLIDE 8-well glass slides (Millipore, catalog number: PEZGS0816)
- Cover glass slides (Fisherbrand, catalog number: 12-545-F)
- 5 ml Serological pipettes (Bio Pioneer, catalog number: GEX0050-S01)
- 10 ml Serological pipettes (Bio Pioneer, catalog number: GEX0100-S01)
- P20 pipette tips sterile with filter
- P200 pipette tips sterile with filter
- 1x Dulbecco’s phosphate buffered saline (DPBS) without calcium and magnesium (Thermo Fisher Scientific, catalog number: 14190250)
- Bovine Serum Albumin (BSA) (Sigma, catalog number: A2153-100G)
- Paraformaldehyde (PFA)
- Tween® 20 (Sigma, catalog number: P9416-100ML)
- Triton X-100 (Manufacturer, catalog number: X-100-500ML)
- Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix (Matrigel) (Corning, catalog number: 354230)
- Rabbit polyclonal anti-ZO-1 antibody (abcam, catalog number: ab59720)
- Mouse monoclonal anti-MiTF antibody (abcam, catalog number: ab12039)
- Mouse monoclonal anti-Bestrophin 1 antibody (Novus Biologicals, catalog number: NB300-164SS)
- Recombinant Rabbit IgG, monoclonal [EPR25A]–Isotype Control (abcam, catalog number: ab172730)
- Mouse IgG1, kappa monoclonal [15-6E10A7]–Isotype Control (abcam, catalog number: ab170190)
- Donkey-anti-Rabbit Alexa FluorTM 647 conjugated antibody (abcam, catalog number: ab150075)
- Goat-anti-Mouse Alexa FluorTM 488 conjugated antibody (Thermo Scientific, catalog number: A-11001)
- ProLong Gold Antifade Reagent with DAPI (Cell Signaling Technologies, catalog number: 8961)
- IF Wash Buffer (see Recipes: Table 12)
- IF Perm Buffer (see Recipes: Table 13)
- IF Staining Buffer (see Recipes: Table 14)
Equipment
- iPSC Cell Culture
- Biosafety cabinet (Labconco, model: Logic+)
- Incubator with humidity and gas control set to maintain 37 °C and 95% humidity in an atmosphere of 5% CO2 in air (Panasonic, model: MCO-170AICUVH-PA)
- Water bath (Thermo Scientific, model: Precision)
- Tissue culture centrifuge with rotors for 15 ml conical tubes and 50 ml conical tubes (Thermo Scientific, model: Legend RT+)
- Phase contrast inverted microscope (objectives: x4, x10, x20) (Olympus, model: CKX41SF)
- (Optional) Phase contrast inverted microscope with camera (objectives: x4, x10, x20) (Thermo Scientific, model: EVOS XL Core)
- Microscope Object marker (Nikon, model MBW10020)
- Pipette aid
- P20 Micropipette
- Freezer -20 °C
- Refrigerator 2-8 °C
- Monolayer plating
- Biosafety cabinet (Labconco, model: Logic+)
- Incubator with humidity and gas control set to maintain 37 °C and 95% humidity in an atmosphere of 5% CO2 in air (Panasonic, model: MCO-170AICUVH-PA)
- Tissue culture centrifuge with rotors for 15 ml conical tubes and 50 ml conical tubes (Thermo Scientific, model: Legend RT+)
- Phase contrast inverted microscope (objectives: x4, x10, x20) (Olympus, model: CKX41SF)
- Phase contrast inverted microscope with camera (objectives: x4, x10, x20) (Thermo Scientific, model: EVOS XL Core)–Optional
- Pipette aid
- P20 Micropipette
- P200 Micropipette
- P1000 Micropipette
- Automated cell counter (Bio-Rad, model: TC20) or a hemocytometer (Hausser Scientific, catalog number: 1483) or equivalent
- Freezer -20 °C
- Refrigerator 2-8 °C
- iPSC-RPE differentiation and cryopreservation
- Biosafety cabinet (Labconco, model: Logic+)
- Incubator with humidity and gas control set to maintain 37 °C and 95% humidity in an atmosphere of 5% CO2 in air (Panasonic, model: MCO-170AICUVH-PA)
- Tissue culture centrifuge with rotors for 15 ml conical tubes and 50 ml conical tubes (Thermo Scientific, model: Legend RT+)
- Phase contrast inverted microscope (objectives: x4, x10, x20) (Olympus, model: CKX41SF)
- (Optional) Phase contrast inverted microscope with camera (objectives: x4, x10, x20) (Thermo Scientific, model: EVOS XL Core)
- Pipette aid
- P20 Micropipette
- P200 Micropipette
- P1000 Micropipette
- Automated cell counter (Bio-Rad, model: TC20) or a hemocytometer (Hausser Scientific, catalog number: 1483) or equivalent.
- Mr. Frosty freezing container (Corning, model: CoolCell® FTS30)
- Refrigerator 2-8 °C
- Freezer -20 °C
- Freezer -80 °C
- Liquid nitrogen vapor tank
- Flow cytometry
- Pipette aid
- P20 Micropipette
- P200 Micropipette
- P1000 Micropipette
- P200 Multichannel micropipette
- Refrigerator 2-8 °C
- Freezer -20 °C
- Flow cytometer (BD Biosciences, model: FACSCanto II) or equivalent
- Immunofluorescence
- Pipette aid
- P20 Micropipette
- P200 Micropipette
- P1000 Micropipette
- Refrigerator 2-8 °C
- Freezer -20 °C
- Confocal laser scanning fluorescence microscope (Olympus, FluoView1000)
Software
- FlowJo (Version 10) (FlowJo, LLC, https://www.flowjo.com/)
- FlowView ASW V03.01.03.03 or V4.2a (Olympus Life Science, https://www.olympus-lifescience.com/en/support/downloads/)
Procedure
- iPSC cell culture
- Thaw iPSC cells
- Prepare 12 ml of mTeSR containing 10 μM ROCK Inhibitor.
- Transfer 9 ml of mTeSR containing 10 μM ROCK Inhibitor into a sterile conical tube labeled with the name of the line.
- Remove vial of cryopreserved cells from liquid nitrogen tank. Keep vial on dry ice.
- Place and shake gently in a 37 °C water bath until a pea-sized ice crystal remains (around 2 min).
- Wipe off excess water from the vial, spray with 70% ethanol before placing in the hood.
- Remove thawed cells from the vial and add gently into 9 ml mTeSR containing 10 μM ROCK Inhibitor in a conical tube. Wash the vial with 1-2 ml of mTeSR containing 10 μM ROCK Inhibitor. Collect all cells in the same conical tube.
- Centrifuge cells for 5 min at 53 x g (500 RPM in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature.
- Aspirate supernatant, and gently resuspend cell pellet in 2 ml of mTeSR containing 10 μM ROCK Inhibitor (1 cryovial is thawed into 1 well of 6-well plate).
- Label a Matrigel plate with name of line, clone and passage number. Aspirate DMEM/F-12 from Matrigel-coated plate. Add +1 to the passage number after thawing.
Note: Do not add +1 to the passage number if the passage number was increased during cryopreservation of iPSCs. - Plate cells resuspended in 2 ml into one well of a Matrigel-coated 6-well plate (final volume 2 ml/well).
- 24 h after plating, observe cells. Wash cells gently with DMEM/F-12 (2 ml/well) to remove cell debris and feed using fresh mTeSR medium without ROCK Inhibitor (2 ml/well).
- Daily, observe the iPSCs, remove the differentiated cells, and change the medium with fresh mTeSR (2 ml/well).
Note: It is critical to maintain iPSC culture differentiation free. - Cells should reach 80-90% of confluency and be ready for passage in about 5 days.
- iPSC passaging using Dispase
- Prepare 1x (2 mg/ml) Dispase solution by adding 9 ml DMEM/F-12 to 1 ml of 10x Dispase (20 mg/ml).
- Allow 1x Dispase solution to come to room temperature.
Note: 1x Dispase solution can be stored at 4 °C for maximum 2 weeks. - Mark any areas of differentiation on the well to be split using the Microscope Object marker.
- Aspirate spent media. Aspirate marked areas of differentiation, if any, by gently tapping a Pasteur pipette within the marked circle. Wash with 2 ml of DMEM/F-12 per well.
- Add 1 ml of 1x Dispase in each well to be split. Incubate at 37 °C for 5 min.
- Check morphology of colonies after 5 min.
When edges of the colonies are slightly curled up, cells are ready to be passaged. If edges of colonies are not curled up, incubate cells at 37 °C for another 1-2 min. Do not incubate with Dispase for longer than 8 min. - Aspirate Dispase from all wells.
- Rinse the wells gently 3 times with DMEM/F12 (2 ml/well).
- Add 1 ml of mTeSR media to each well to be passaged.
- Use a glass serological pipette to detach colonies. Hold the pipette at a 90° angle to the surface of the plate. Scrape across the surface of the 6-well plate in the motion outlined in Figure 1 (start from top left side of the well and zig-zag tightly down to bottom-right side, then turn plate clockwise or counterclockwise and scrape again). Scrape until at least 90% of the colonies are detached from the well.
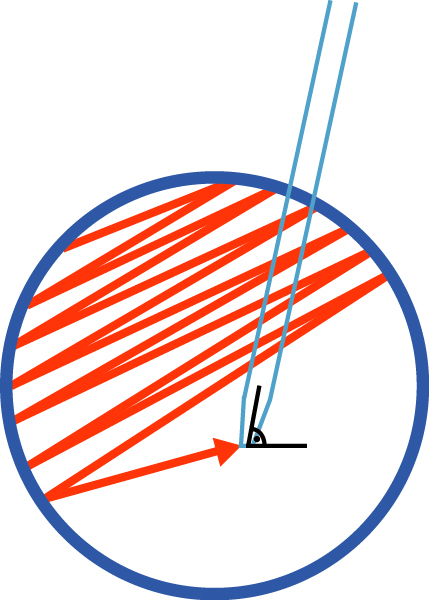
Figure 1. Pattern of movement of a glass serological pipette during the iPSCs passaging with Dispase. After scraping the well in one direction, turn the plate clockwise or counterclockwise by 90° and scrape remaining iPSC colonies again. About 90% cells should be detached from the well. Please refer to the section on iPSC passaging using Dispase for details. - Wash plate with the volume of mTeSR required to bring cells up to the final volume needed to seed a new Matrigel-coated vessel. Calculate the final volume considering 2 ml per each well to be seeded with passaged cells. For example, if cells are to be passaged 1 to 3 the final volume will be 6 ml, therefore the volume of mTeSR used to wash the plate is 5 ml.
- Seed cells on a new Matrigel plate plating 1 ml per well and then add 1 ml more. Plate cells dropwise across the entire surface of the well to ensure uniform plating.
- Observe seeded cells under microscope to ensure even plating.
- Place in a 37 °C incubator. Shake the plate in T-shape to homogenously distribute the colonies pieces in the well.
- Twenty-four hours after plating gently, wash cells with DMEM/F-12 before adding fresh mTeSR medium.
Note: For a healthy and efficient iPSC culture, it is critical to plate cells uniformly. Plate cells uniformly across the entire surface of the well and, when plating multiple well,s plate cells uniformly across all wells.
- Thaw iPSC cells
- Monolayer plating
Note: After thawing an iPSC line, passage cells with Dispase at least once before plating monolayer.- Remove 6-well plates from the incubator. When iPSC cells are at around 80% confluency (cells are ready for a passage), iPSC cells are ready for Monolayer. Mark all differentiated cells, which need to be removed.
- Aspirate the spent medium. Remove all marked differentiated cells and wash cells with DMEM/F-12 (2 ml/well).
- Aspirate DMEM/F-12 and add 1 ml of room temperature Accutase to well of a 6-well plate. Incubate cell for 8 min at 37 °C.
- After 8 min of incubation, add 1 ml per well of mTeSR containing 5 μM ROCK inhibitor and re-suspend cells as single cells without scraping plate surface, using a P1000 pipette. Pipette cells 10-12 times, turn the dish by 180° (upside down) and pipette 5 more times. Collect cells from all wells in a 50 ml conical tube. You should not see any cell clumps.
- Wash all wells twice with 5 ml of mTeSR containing 5 μM ROCK Inhibitor. Collect all cells in the same conical tube.
- Centrifuge the cells at 53 x g (500 RPM in a Sorvall 75006445 rotor with 75006441 K buckets) for 7 min at room temperature. Aspirate the supernatant and resuspend cells in 10 ml of mTeSR containing ROCK Inhibitor.
- Mix the pooled cell suspension by inverting 20 times or more if necessary. Perform the live cell count using 0.4% Trypan Blue Solution.
Note: iPSC cell viability should be not lower than 80%. - Prepare required number of cells. Optimal cell number will vary depending on the scale of differentiation. iPSC-RPE differentiation protocol requires 2.5 x 104 live cells per cm2 (2 x 106 per one 100 mm dish). For one 100 mm dish, prepare in a 15 ml conical tube 11 ml of cell suspension containing 2.2 x 106 cells. Mix cell suspension very well by inverting the tube 20 times.
- Add 10 ml of cell suspension per 100 mm dish dropwise using a 10 ml pipette.
e (T-shape).
- Place plates in the incubator without stacking the plates. Incubate the cells until next morning, at 37 °C, 5% CO2.
- Next day change medium for fresh mTeSR without ROCK inhibitor (10 ml/100 mm dish). Monolayer for iPSC-RPE cell differentiation requires culturing cells for about 4-5 days until the monolayer reaches 80% confluency. Change medium with fresh mTeSR daily.
- iPSC-RPE differentiation
Refer to Figure 2 for a schematic representation of the differentiation protocol.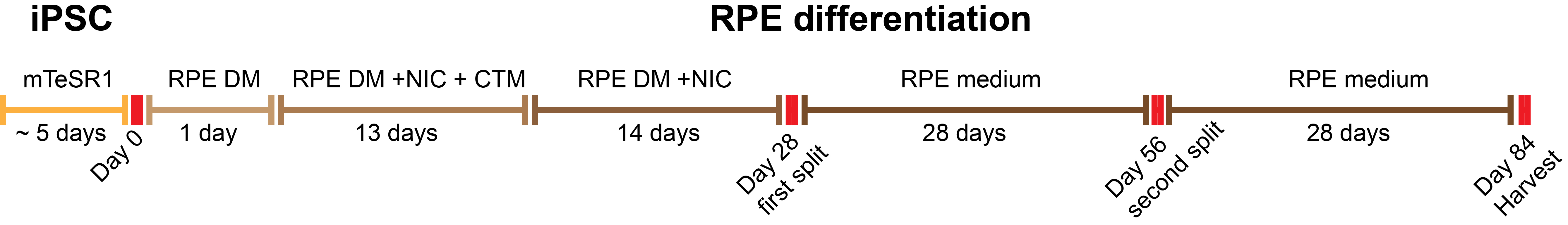
Figure 2. Schematic representation of the iPSC-RPE differentiation protocol. Nicotinamide (NIC), Chetomin (CTM). iPSC monolayer is cultured until the cells reach 80% confluency. iPSC-RPE differentiation is initiated and driven using small molecules (NIC and CTM). iPSC-RPEs continue to differentiate and expand for a total of 12 weeks. Whole culture passages promote expansion of iPSC-RPE cells. Adapted from Smith et al., 2019.- Day 0 (D0)–When iPSC monolayer reaches about 80% confluency initiate the iPSC-RPE differentiation by replacing mTeSR medium with RPE DM medium (see Recipes: Table 5) (24 ml/100 mm dish)
- D1–24 h after initiation of the differentiation replace spent RPE DM medium with fresh RPE DM medium supplemented with 10 mM Nicotinamide (see Recipes: Table 9) and 50 nM Chetomin (see Recipes: Table 8) (24 ml/100 mm dish).
- Daily change spent medium with fresh RPE DM medium supplemented with 10 mM Nicotinamide (NIC) and 50 nM Chetomin (CTM) (24 ml/10 cm dish).
Note: Due to very high cell number, high proliferative rate and metabolic activity of iPSC-RPE cells to obtain a healthy and robust iPSC culture it is strongly recommended to maintain the schedule of 24 h media change throughout the entire differentiation. - On D14 change spent medium with fresh RPE DM medium supplemented with 10 mM Nicotinamide (24 ml/100 mm dish).
- Daily change spent medium with fresh RPE DM medium supplemented with 10 mM Nicotinamide (24 ml/100 mm dish).
Note: First cells start to acquire melanin pigmentation and the characteristic polygonal shape after 2-3 weeks. - On D28 passage the cells–first passage.
- (Optional) Take images of the cells–(Figure 3-D28-left panel).
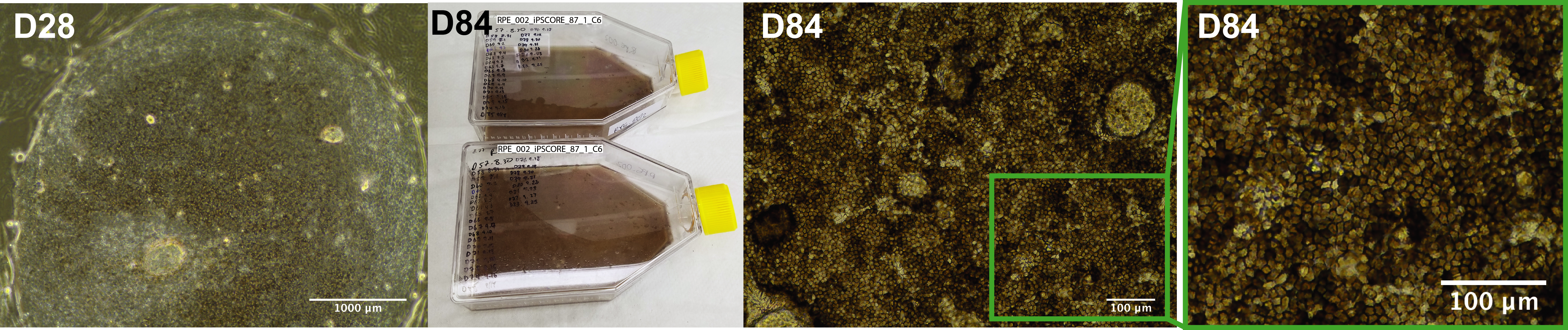
Figure 3. Images of the iPSC-RPE cells. Bright-field image of iPSC-RPE at Day 28 (D28) of the differentiation iPSC-RPE cells appear as small clusters of polygonal pigmented cells which expand over time (left); T150 flasks containing iPSC-RPE cells (iPSCORE 87_1) at Day 84 (D84) (middle left); Bright-field image of iPSC-RPE sample (iPSCORE_42_1) at Day 84 illustrating a highly organized monolayer with strong melanin pigmentation (middle right) and characteristic polygonal shape (right). Adapted from Smith et al., 2019. - Aspirate spent medium and wash cells with PBS (10 ml/100 mm dish).
- Aspirate PBS and add 5 ml of room temperature Accutase to a 100 mm dish. Incubate cells for 12 min at 37 °C.
- After 12 min of incubation add 5 ml per dish of RPE DM medium, re-suspend cells in Accutase as single cells without scraping the surface of the plate, using a P1000 pipette. If cells are difficult to remove from a dish, use a cell scraper.
- Collect cells in a 50 ml conical tube.
- Wash plate three additional times, each with 5 ml of RPE DM medium. Collect all cells in the same conical tube.
- Centrifuge cells for 8 min at 136 x g (800 RPE in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature.
- After centrifugation, aspirate the supernatant and resuspend the cells in 10 ml of RPE medium (see Recipes: Table 6).
- Gently pass all cells through a 70 μm strainer. Add medium for a total of 20 ml of final volume passing it through the strainer.
- Plate cells on two fresh 100 mm dishes which were coated overnight with Matrigel.
- Using a 10 ml pipette add dropwise 10 ml of cell suspension per 100 mm dish.
Note: It is critical to plate cells uniformly on the entire surface of the plates. To help distribute the cells uniformly plate one dish at the time and shake the newly plated dish in a cross shape (T-shape).
Optional: Instead of using two 100 mm dishes plate cells on one T150 flask. - Place plates in the incubator. Incubate the cells until next day, at 37 °C, 5% CO2 without stacking the plates.
- (Optional) Take images of the cells–(Figure 3-D28-left panel).
- Twenty-four hours after split, change medium for fresh RPE medium (10 ml/100 mm dish). If using T150 flask use 25 ml/T150 flask.
- Change medium daily for fresh RPE medium (10 ml/100 mm dish) for 4 weeks. If using T150 flask use 25 ml/T150 flask.
- On D56 passage the cells–second passage.
Perform the passage following Steps C6a-C6g.
Note: If cells were plated on a T150 flask, incubate cells with 10 ml of Accutase per flask and use a cell scraper to recover all the cells. To collect all the cells, wash flask three times, each time with 10 ml of medium.- After centrifugation, aspirate the supernatant and resuspend the cells in 20 ml of RPE medium.
- Gently pass all cells through a 70 μm strainer. Add medium for a total of 50 ml of final volume passing at least 20 ml of it through the strainer.
- Plate cells onto two fresh T150 flasks which were coated overnight with Matrigel.
- Add 25 ml of cell suspension per each T150 flask dropwise using a 10 ml pipette.
Note: It is critical to plate cells uniformly on the entire surface of the plates which were coated overnight with Matrigel. To help distribute the cells uniformly plate one flask at the time and shake the newly plated flasks in a cross shape (T-shape). - Place flasks in the incubator without stacking. Incubate the cells until next morning, at 37 °C, 5% CO2.
- Twenty-four hours after split, change medium with fresh RPE medium (25 ml/T150 flask).
- Change medium daily with fresh RPE medium (25 ml/T150 flask) for 4 weeks.
- On D84 collect all cells.
- (Optional) Take images of the cells. Refer to Figure 3 for an example of expected iPSC-RPE yield, pigmentation (Figure 3–panel middle left and middle right) and cell morphology (Figure 3–panel middle right and right).
- Aspirate spent medium and wash cells with PBS (20 ml/T150 flask).
- Aspirate PBS and add 10 ml of room temperature Accutase to each flask. Incubate for 12 min at 37 °C.
- After 12 min of incubation, add 10 ml per well of RPE medium re-suspend cells. Use cell scraper to remove all cells.
- Collect cells in a 50 ml conical tube.
- Wash plate three additional times with 10 ml of RPE medium. Collect all cells in the same conical tube.
- Centrifuge cells for 8 min at 136 x g (800 RPE in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature.
- After centrifugation, aspirate the supernatant and resuspend the cells in 20 ml of RPE medium.
- Gently pass all cells through a 70 μm strainer. Add medium for a total of 40 ml of final volume passing at least 20 ml of it through the strainer.
- Mix the cell suspension by inverting 20-30 times. Perform the live cell count using 0.4% Trypan Blue Solution.
- When performing the live cell count of iPSC-RPE using automated cell counter, cell viability may be inaccurately scored as high due to the high melanin concentration in the cells. Depending on the experimental needs, iPSC-RPE cells can be cryopreserved for future experiments, fixed for flow cytometry analysis and/or plated for immunofluorescence analysis.
- Cells start to acquire melanin pigmentation and the characteristic polygonal shape after 2-3 weeks in culture and about 1 week after each passage. The first signs of the cells starting to acquire the pigmentation is slightly grayish (“dirty”) hue of the color of the medium. When cells are strongly pigmented the medium also acquires dark color. Cell suspension of concentrated iPSC-RPE at D84 appears black, similarly to the pelleted iPSC-RPE-cells which are also black (Figure 4).

Figure 4. Images of the iPSC-RPE cells at D84. iPSC-RPE cells (iPSCORE_29_1) collected prior to the centrifugation (left) and after centrifugation (right).
- Cryopreservation of iPSC-RPE
- Prepare 2x iPSC-RPE freezing medium by preparing 20% DMSO solution in FBS. Prepare 0.25 ml of 2x iPSC-RPE freezing medium per each cryovial intended to be cryopreserved. Freeze cells at a final density of 1.2 x 107/ml (depending on the downstream experiments the volume and the concentration of the cryopreserved iPSC-RPE cells in a single cryovial can be modified).
Optional: If a serum free conditions are required, prepare the 2x iPSC-RPE freezing medium using KOSR instead of FBS. - Prepare and print the labels for cryovials. Prepare n + 2 number of labels (n = number of cryovials to be cryopreserved). Prepare and affix the labels on all cryovials to be frozen, use one label for the Mr. Frosty and one label for record keeping (i.e., lab book).
- After the live cell count (Step C12j), determine how many cells should be cryopreserved and transfer desired number of cells into a new 15 ml or 50 ml conical tube.
- Centrifuge cells for 5-8 min at 136 x g (800 RPM in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature (adjust the time of centrifugation depending on the volume of cells).
- After centrifugation, aspirate the supernatant and resuspend the cells in 0.25 ml of FBS (or KOSR) per each cryovial to be frozen at the concentration of 2.4 x 107/ml (i.e., for 10 cryopreserved vials resuspend 6 x 107 cells in 2.5 ml of FBS or KOSR).
- Open all pre-labeled cryovials and add 0.25 ml of cell suspension to each cryovial.
- Add 0.25 ml of 2x iPSC-RPE freezing medium to each cryovial containing the iPSC-RPE cell suspension.
- Close all cryovials and gently invert them 5-6 times to mix cell suspension and 2x iPSC-RPE freezing medium. Transfer cryovials to Mr. Frosty freezing container.
- Immediately transfer Mr. Frosty into a -80 °C freezer. When freezing large number of cryovials (i.e., multiple Mr. Frosties) prepare individual batches, with each batch containing only the number of cryovials that will fit into one Mr. Frosty.
- After 24-48 h, transfer the cells into a liquid nitrogen vapor tank. Update accordingly the records (i.e., box maps).
- Prepare 2x iPSC-RPE freezing medium by preparing 20% DMSO solution in FBS. Prepare 0.25 ml of 2x iPSC-RPE freezing medium per each cryovial intended to be cryopreserved. Freeze cells at a final density of 1.2 x 107/ml (depending on the downstream experiments the volume and the concentration of the cryopreserved iPSC-RPE cells in a single cryovial can be modified).
- Flow cytometry
- After the live cell count (Step C12j), determine how many cells should be fixed for flow cytometry analysis and transfer desired number of cells into a 15 ml conical tube. Use at least 2-5 x 106 cells.
- Centrifuge cells for 5 min at 136 x g (800 RPM in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature.
- After centrifugation, aspirate the supernatant and resuspend the cells in 10 ml of PBS.
Optional: If the volume of cell suspension used for flow cytometry in smaller than 0.5 ml then add directly to the cells 14 ml of PBS and centrifuge mix of cells and PBS for 8 min at 136 x g (800 rpm in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature. - Fix and permeabilize iPSC-RPE cells using the Fixation/Permeabilization Solution Kit with BD GolgiStopTM following manufacturer recommendations. After the last centrifugation, aspirate the supernatant and resuspend the cells in the 1x BD Perm/WashTM Buffer at the concentration of 1 x 107/ml. For each flow cytometry staining use 2.5 x 105 cells.
Note: Staining of 2.5 x 105 cells allows for an efficient cells and reagent usage, however it is also possible to use 1 x 106 cells per staining maintaining the same antibodies dilution ratios. - Transfer 25 μl of fixed and permeabilized cells into 5 wells of a 96-well round bottom assay plate. In order to limit usage of the antibodies and cells when staining multiple lines, mix equal number of cells from each line and transfer 25 μl of the cell mix into four control wells [Recombinant Rabbit IgG, monoclonal class control antibody (Rb-IgG), Mouse IgG1, kappa monoclonal (M-IgG1), anti ZO-1 antibody (ZO1) and anti MiTF antibody (MiTF)].
- Following Table 15 add appropriate concentrations of the antibodies in each well. Using a multichannel pipette set for 20 μl mix cells and antibodies gently by pipetting up and down 20 times.
- Incubate cells with primary antibodies for 1 h at room temperature.
- After 1 h, add 150 μl of FACS buffer (see Recipes: Table 10).
- Centrifuge plate at 863 x g (2,000 RPM in a Sorvall 75006445 rotor with 75006441 K buckets) for 8-10 s counting from when the speed reaches 863 x g (2,000 rpm in a Sorvall 75006445 rotor with 75006441 K buckets) at room temperature.
In detail:- Set the centrifuge for 863 x g (2,000 RPM), 1 min, room temperature); start the centrifuge and wait until the speed reaches 863 x g (2,000 rpm).
- Count to 8-10 s and stop the centrifuge.) The pellet after the centrifugation should be clearly visible especially when using 1 x 106 cells.
- After centrifugation, gently aspirate the supernatant, being very careful not to aspirate any cells. If using vacuum to aspirate cells, use a P20 tip without a filter (or a P200 + P20 tips without filters). Leave about 20 μl of liquid in each well to avoid aspirating the cells.
- Using a multichannel pipette add 200 μl of FACS buffer and mix cells gently 5-6 times.
- Centrifuge plate like in Step E9.
- Repeat Steps E10-E12 to wash the cells one more time.
- Resuspend cells in 50 μl of 1x BD Perm/WashTM Buffer.
- Following Table 15 add appropriate concentrations of the antibodies in each well. Using a multichannel pipette set for 40 μl mix cells and antibodies gently by pipetting up and down 20 times.
- Incubate cells with secondary antibodies for 45 min at room temperature in darkness.
- After 45 min repeat Steps E8-E13.
- After the last centrifugation aspirate the supernatant and resuspend the cells in 200 μl of FACS-FIX Buffer (see Recipes: Table 11). Using a multichannel pipette resuspend the cells by pipetting 5-6 times.
- Using a P1000 pipette transfer each sample, one at a time, into a CorningTM FalconTM Test Tubes with Cell Strainer Snap Cap passing the cells through the strainer in a cap.
- With an additional 250 μl wash each well and transfer to the appropriate tube passing the cells through the strainer in a cap. Depending on the number of cells used for staining dilute the cells to an appropriate concentration to avoid clogging the flow cytometer.
- Place all the tubes in an appropriate rack and wrap them in an aluminum foil to protect from light.
- Proceed with acquisition using a flow cytometer FACS Canto II (or alternative flow cytometer).
- Perform the flow cytometry analysis using FlowJo software V 10.4. Refer to the Figure 5 for an example of iPSC-RPE flow cytometry staining results.
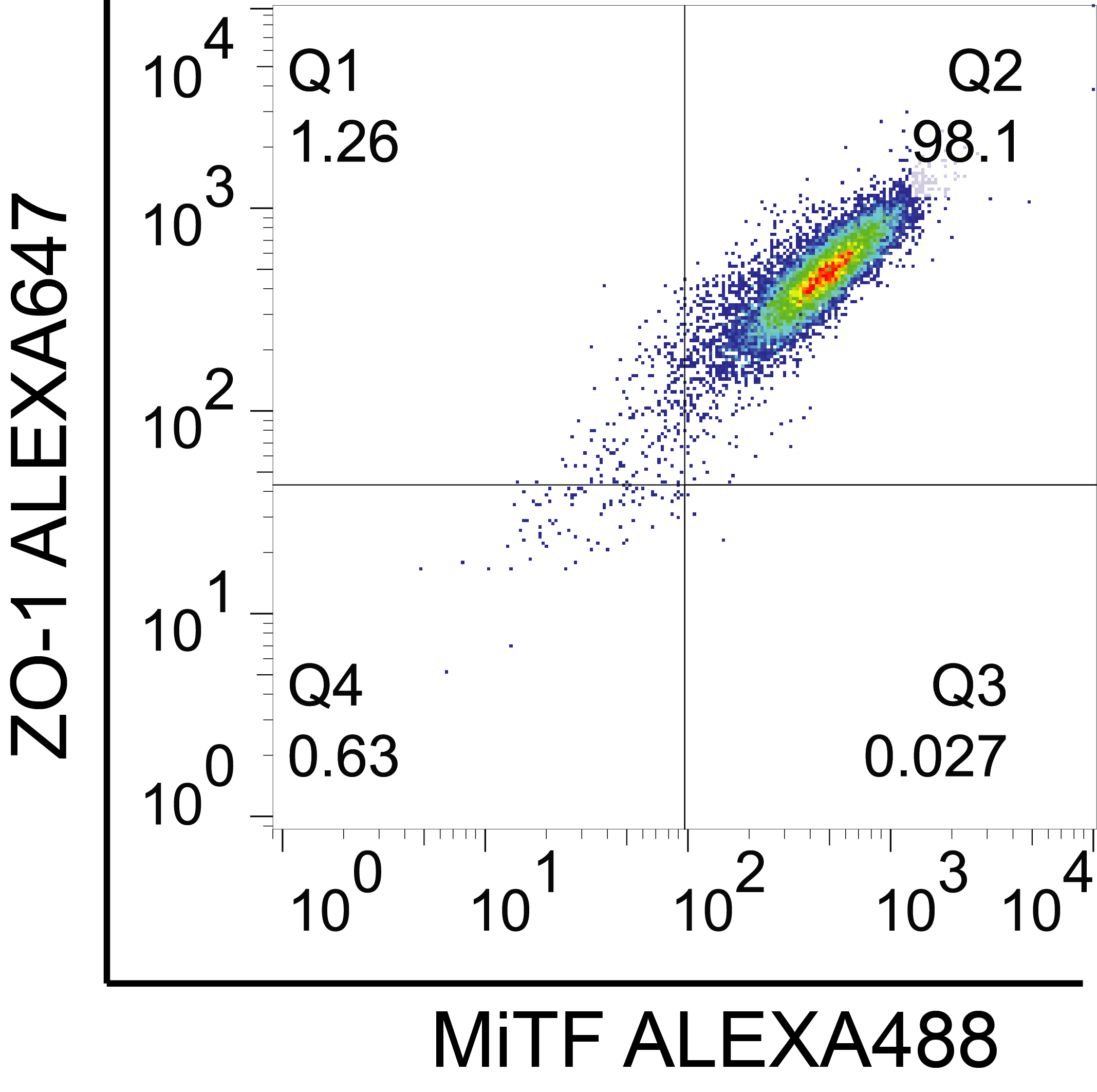
Figure 5. Flow-cytometry analysis of iPSC-RPE (iPSCORE_42_1) at Day 84 showing high co-staining of Zonula Occludens 1 (ZO-1) and Microphthalmia-associated Transcription Factor (MiTF). Adapted from Smith et al., 2019.
- Immunofluorescence
- Coat Millicell EZ SLIDE 8-well glass slides overnight with Matrigel.
- Plate fresh or cryopreserved iPSC-RPE cells on the Matrigel coated Millicell EZ SLIDE 8-well glass slides. Plate at least 6 wells per line at the density of 1-1.5 x 106/cm2.
- Culture cells for 10 days until they reach full confluency, re-acquire polygonal shape and pigmentation.
- Aspirate the medium and wash cells twice with PBS. Aspirate the PBS.
- Fix cells with 4% PFA for 10 min at room temperature.
- Remove the PFA solution and wash cells twice with freshly prepared IF Wash Buffer (see Recipes: Table 12). Aspirate the IF Wash Buffer.
- Saturate and permeabilize the cells using IF Perm Buffer (see Recipes: Table 13). Incubate the cells for 20 min at room temperature.
- In the last 5 min of the saturation and permeabilization prepare the primary antibody solutions in IF Staining Buffer (see Recipes: Table 14 and Table 15) for the appropriate concentrations of the antibodies. Store antibodies solutions on ice until use.
- After saturation and permeabilization aspirate all the buffer and add antibodies solutions to the appropriate wells.
- Incubate cells with the antibodies solution overnight at 4 °C.
- Next day (morning) prepare the secondary antibody solutions in IF staining Buffer. Refer to the Table 15 for the appropriate concentrations of the antibodies. Keep antibodies solutions on ice until use, protected from light.
- Aspirate the primary antibodies solutions and wash cells three times with PBS. After last wash aspirate all PBS.
- Immediately add the secondary antibodies solutions to the appropriate wells. Incubate cells for 1 h at room temperature in darkness.
- Aspirate the secondary antibodies solutions and wash cells three times with PBS. After last wash aspirate all PBS.
- Detach the walls of the Millicell EZ SLIDE 8-well glass slides.
- Add ProLong Gold Antifade Reagent with DAPI following manufacturer’s recommendations and gently mount the cover glass slide avoiding bubbles. Use a pencil rubber to gently remove any bubbles. Store the slide(s) at room temperature for several hours (best until next day) in darkness to allow proper mounting.
- Acquire images using an appropriate immunofluorescence microscope (best is a confocal laser scanning fluorescence microscope). Refer to Figure 6 for an example of iPSC-RPE immunofluorescence staining.
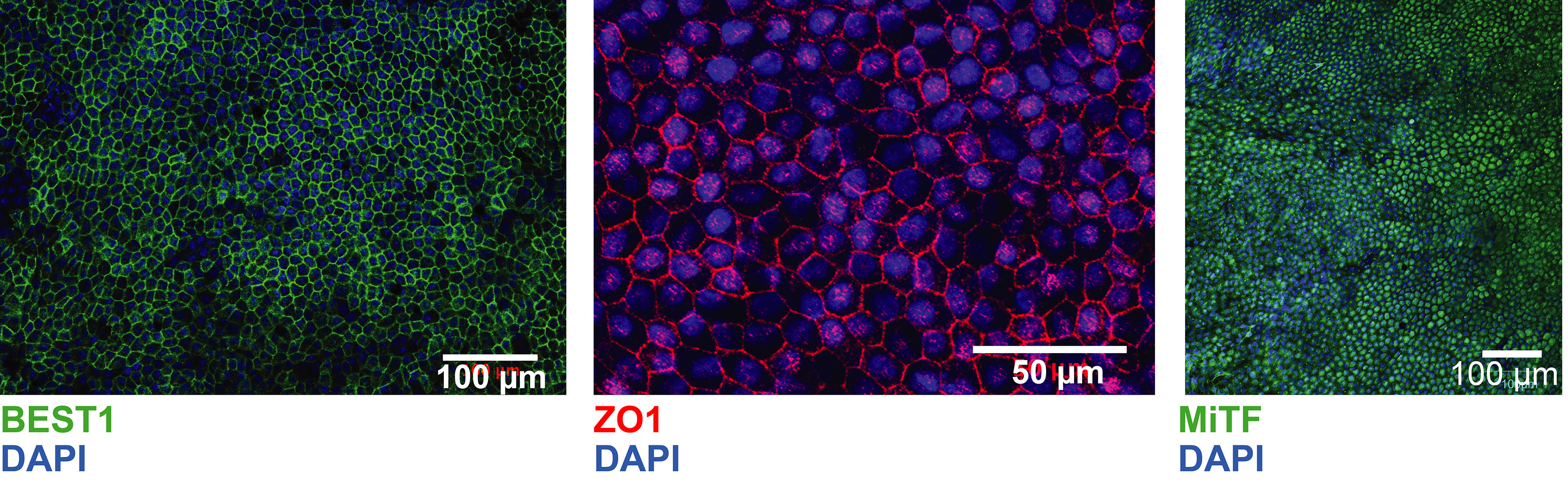
Figure 6. Immunofluorescence analysis of Bestrofin 1 (BEST1) (iPSCORE_29_1), ZO-1 (iPSCORE_29_1), and MiTF (iPSCORE_42_1). ZO-1 appears as sharp cell membrane staining; BEST1 membrane staining appears “fuzzier” compared to ZO1 staining; MiTF nuclear staining. Adapted from Smith et al., 2019.
Recipes
- Cell culture reagents and media preparation
Table 1. Preparation of Matrigel solution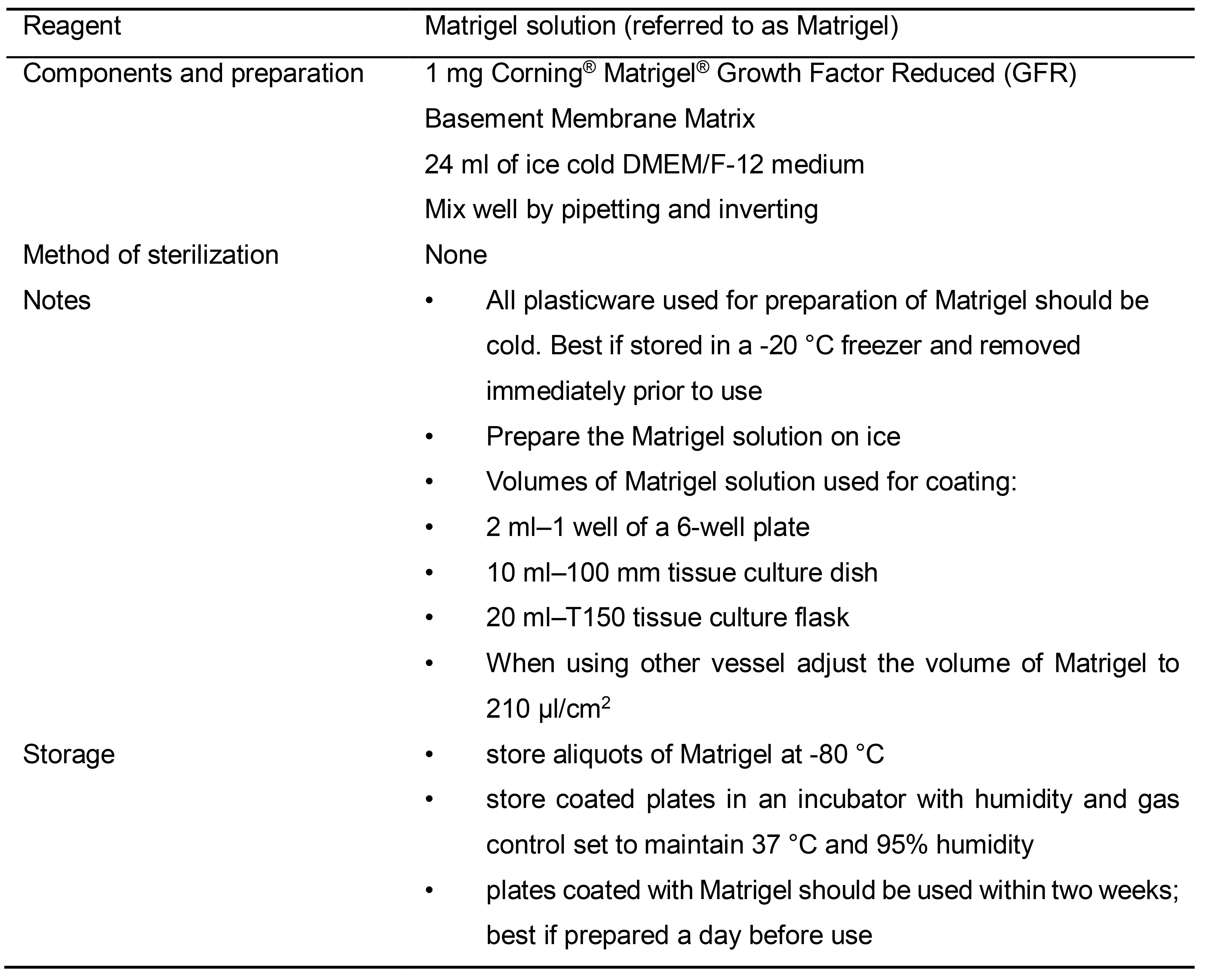
Table 2. Preparation of 1.10 mM ROCK inhibitor, Y-27632 dihydrochloride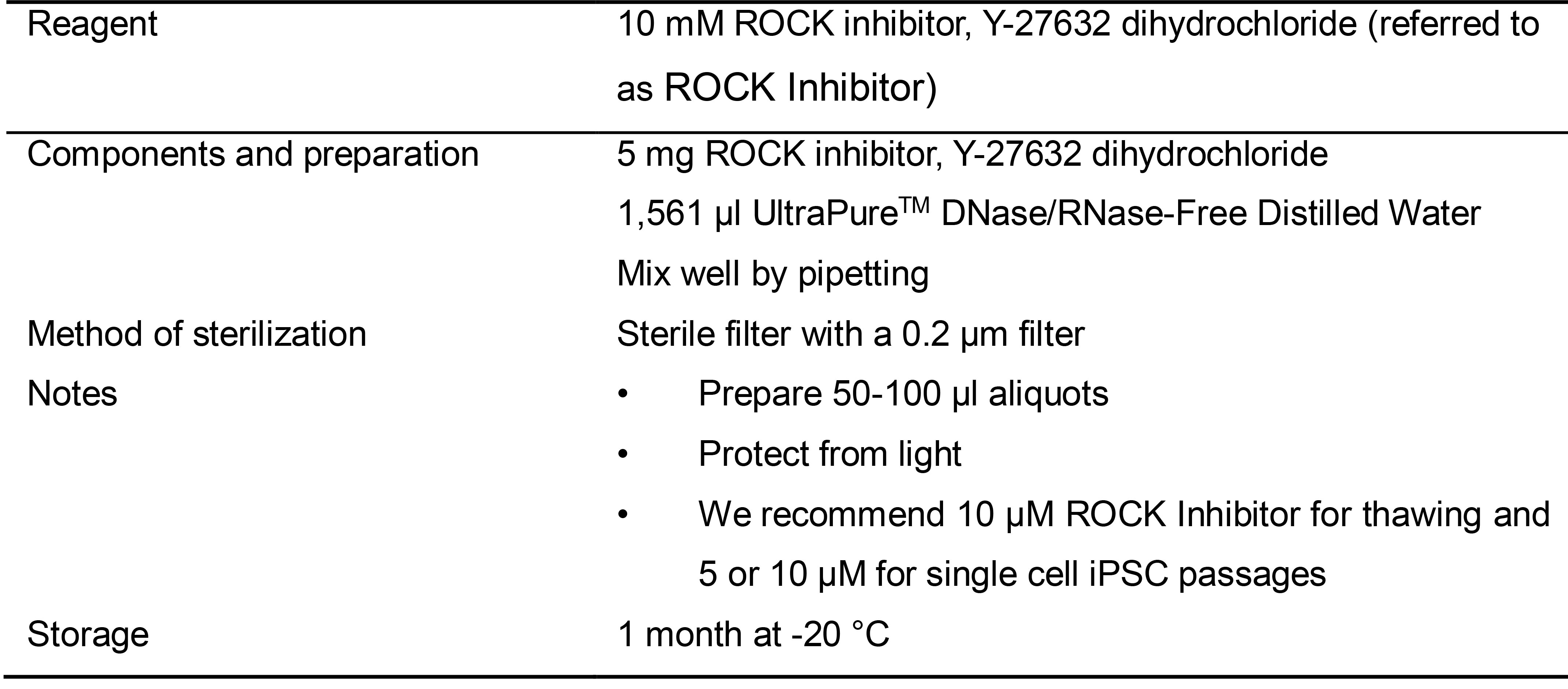
Table 3. Preparation of 10x Dispase Solution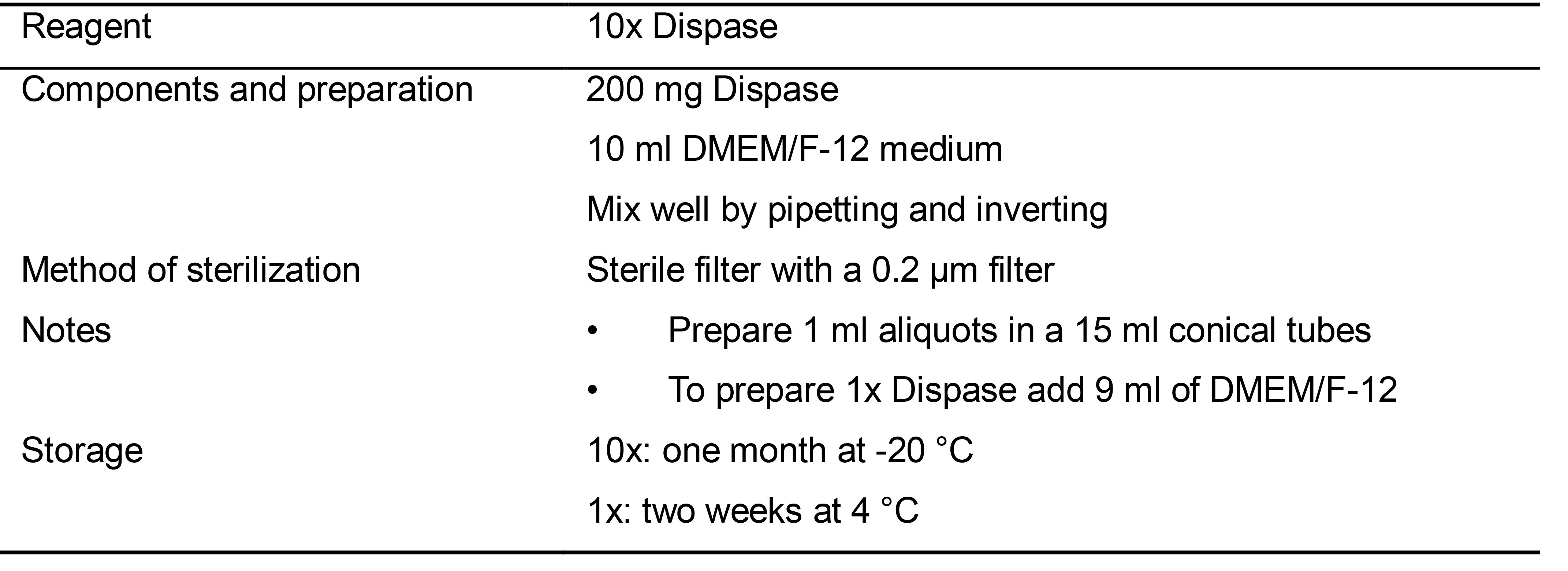
Table 4. Preparation of mTeSRTM1 complete medium
Table 5. Preparation of RPE DM medium (following Maruotti et al., 2015)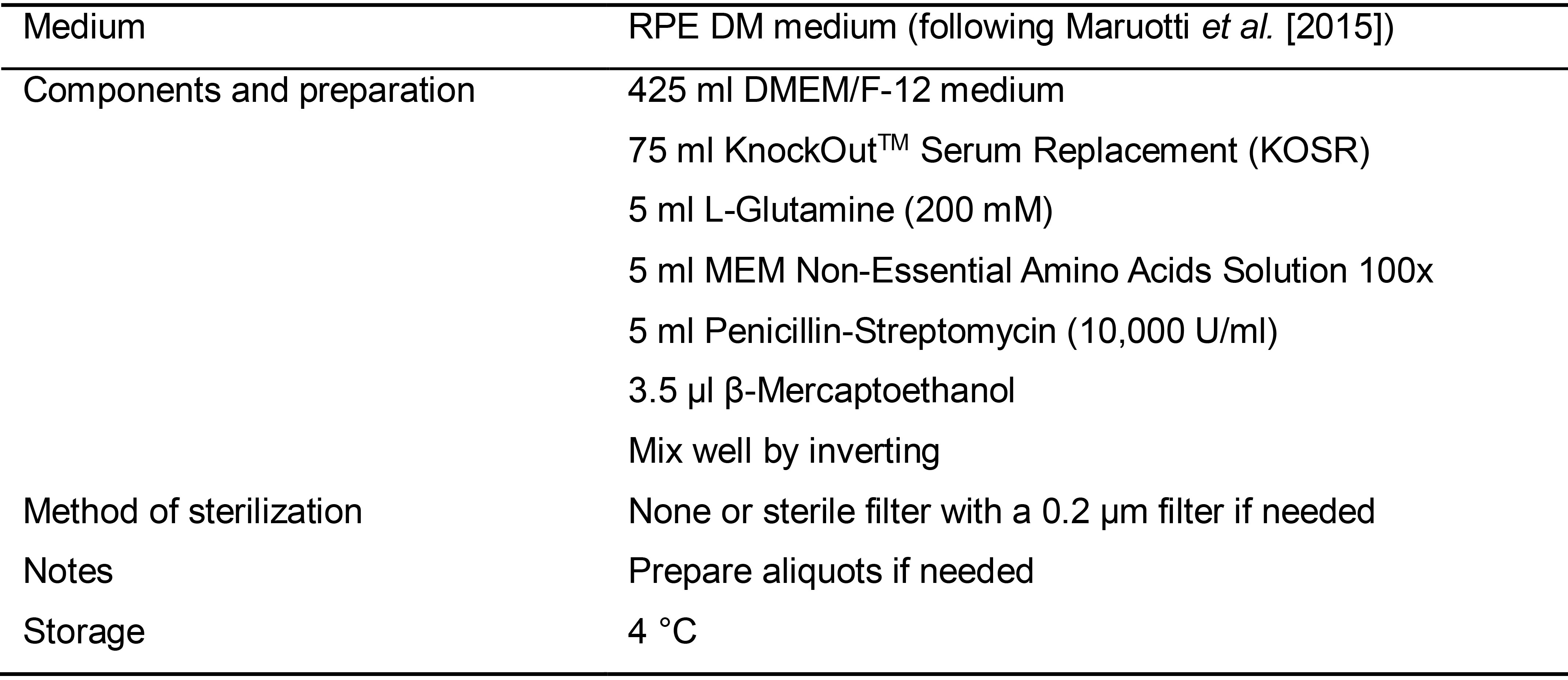
Table 6. Preparation of RPE medium (following Maruotti et al., 2015)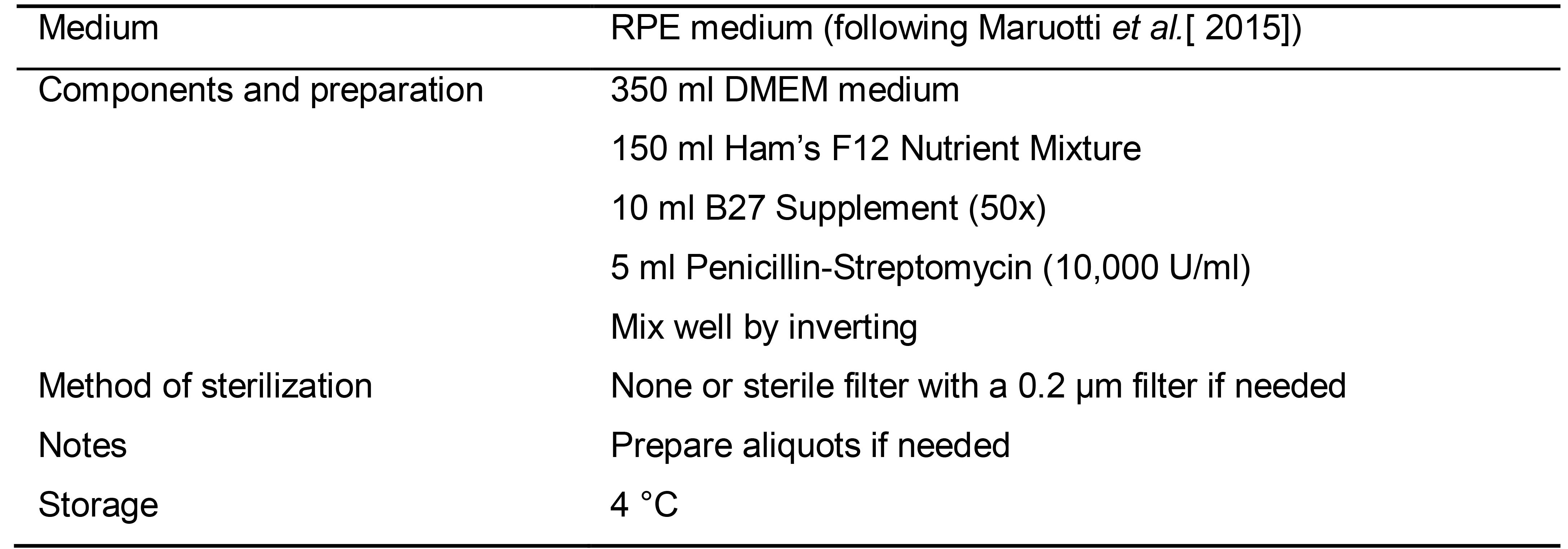
Table 7. Preparation of 2x iPSC-RPE freezing medium (for 5 ml)
Table 8. Preparation of 1 mM Chetomin solution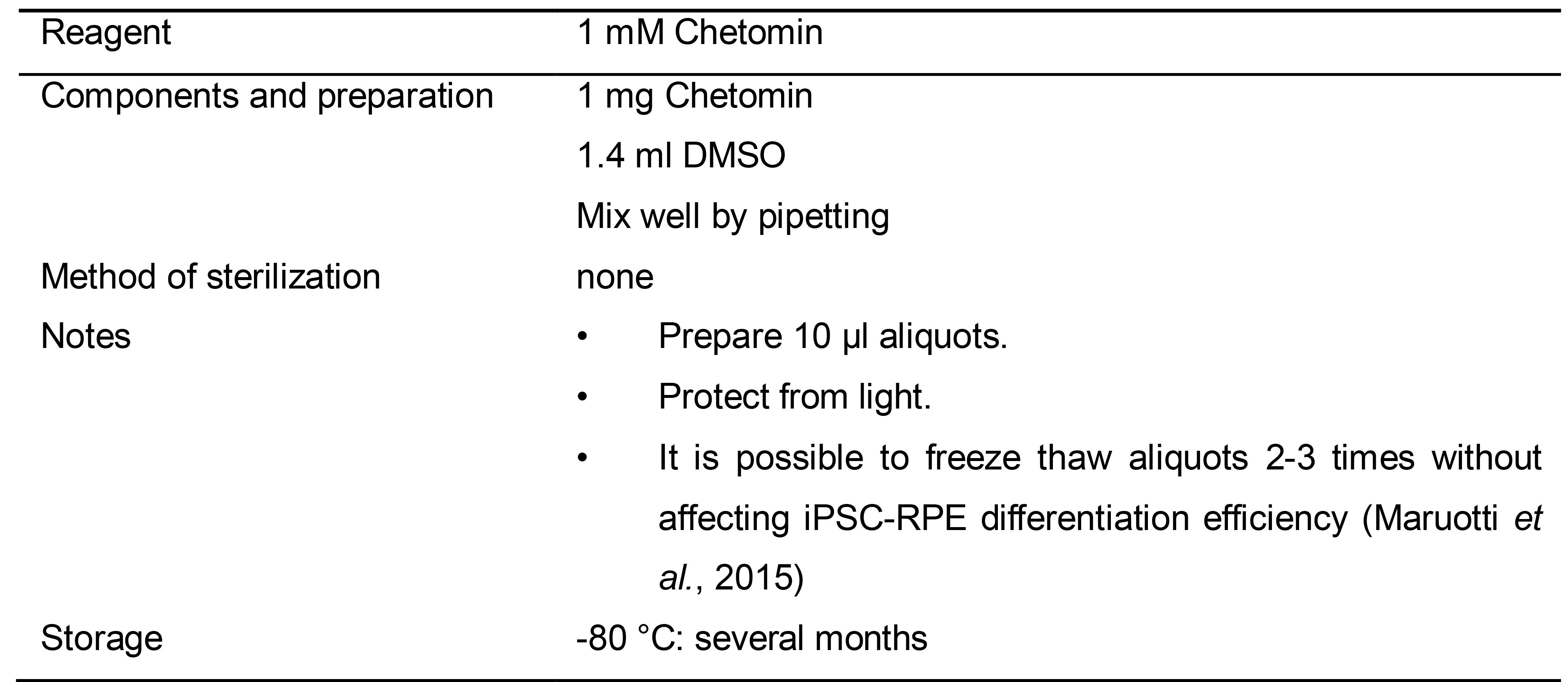
Table 9. Preparation of 1M Nicotinamide (100x) solution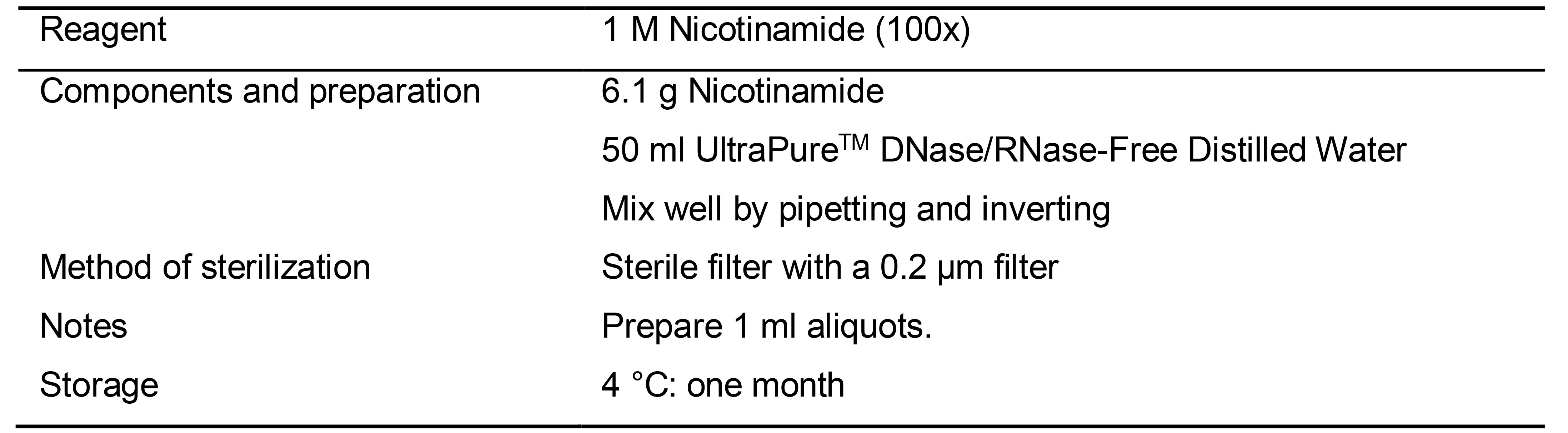
- Buffer preparation
Table 10. Preparation of FACS Buffer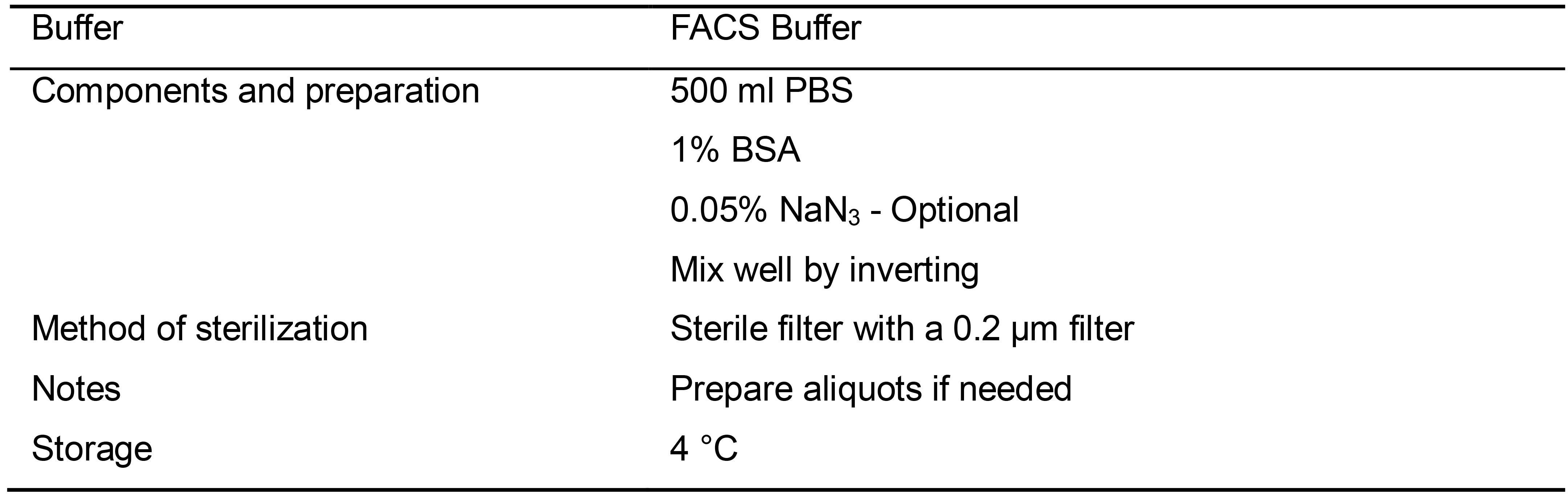
Table 11. Preparation of FACS-FIX Buffer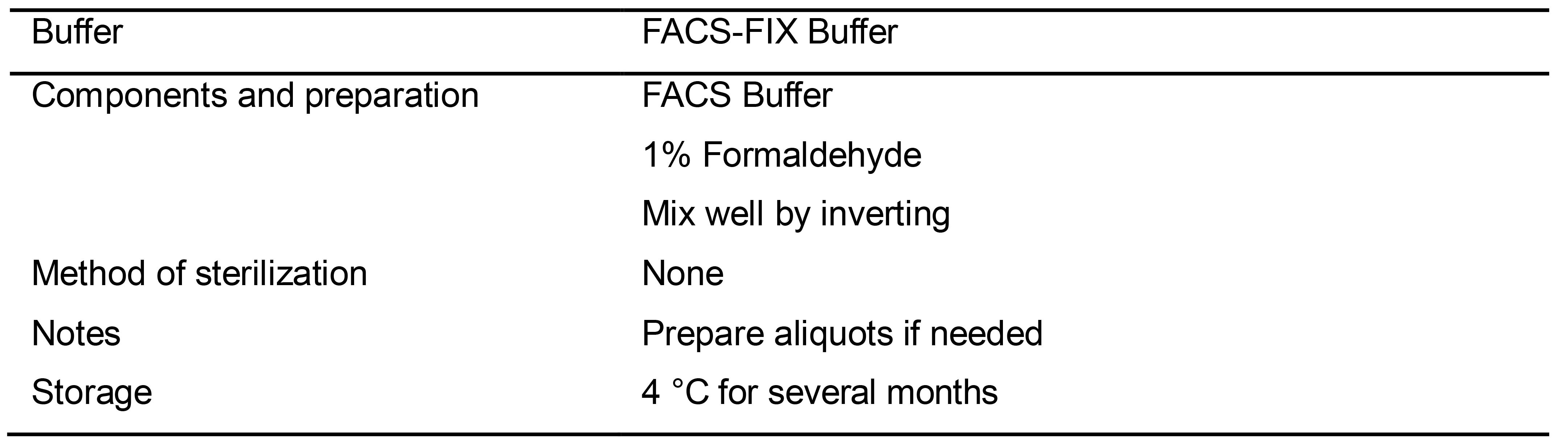
Table 12. Preparation of IF Wash Buffer
Table 13. Preparation of IF Perm Buffer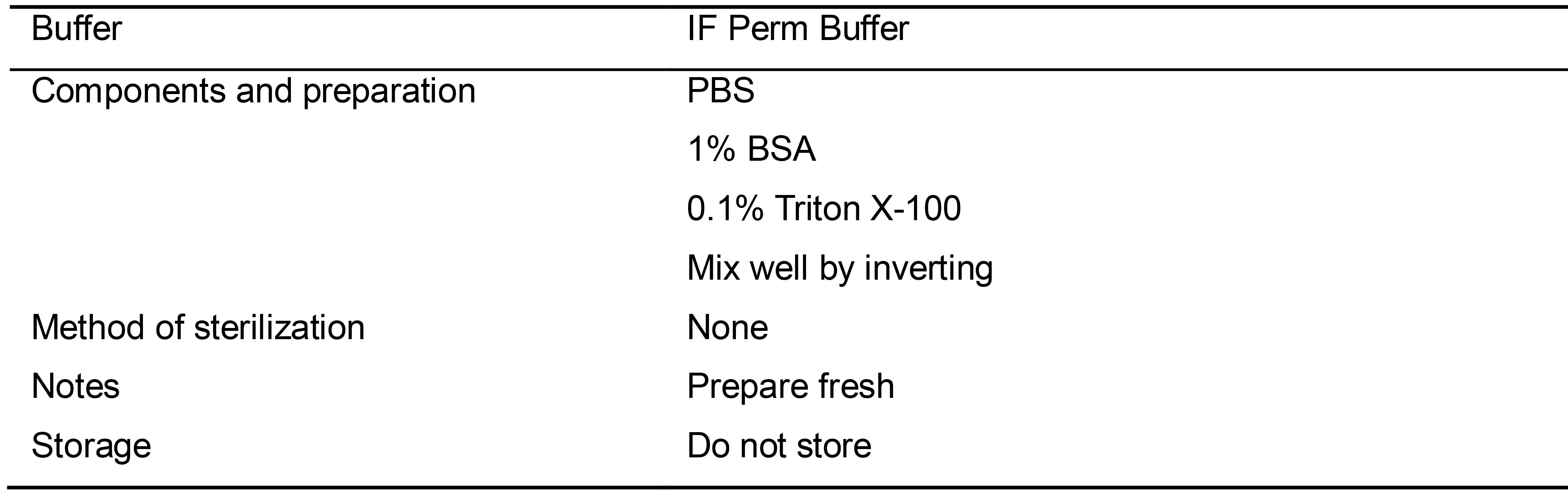
Table 14. Preparation of IF Staining Buffer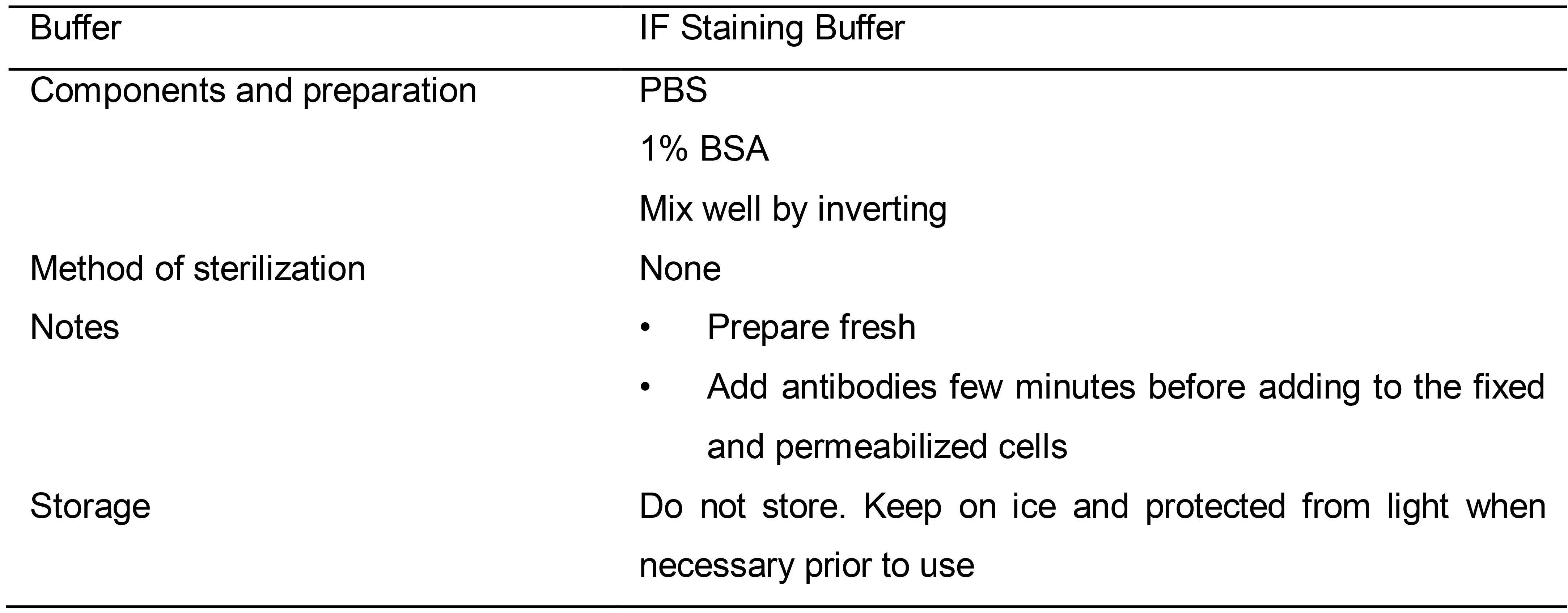
- Antibodies
Table 15. Antibodies concentrations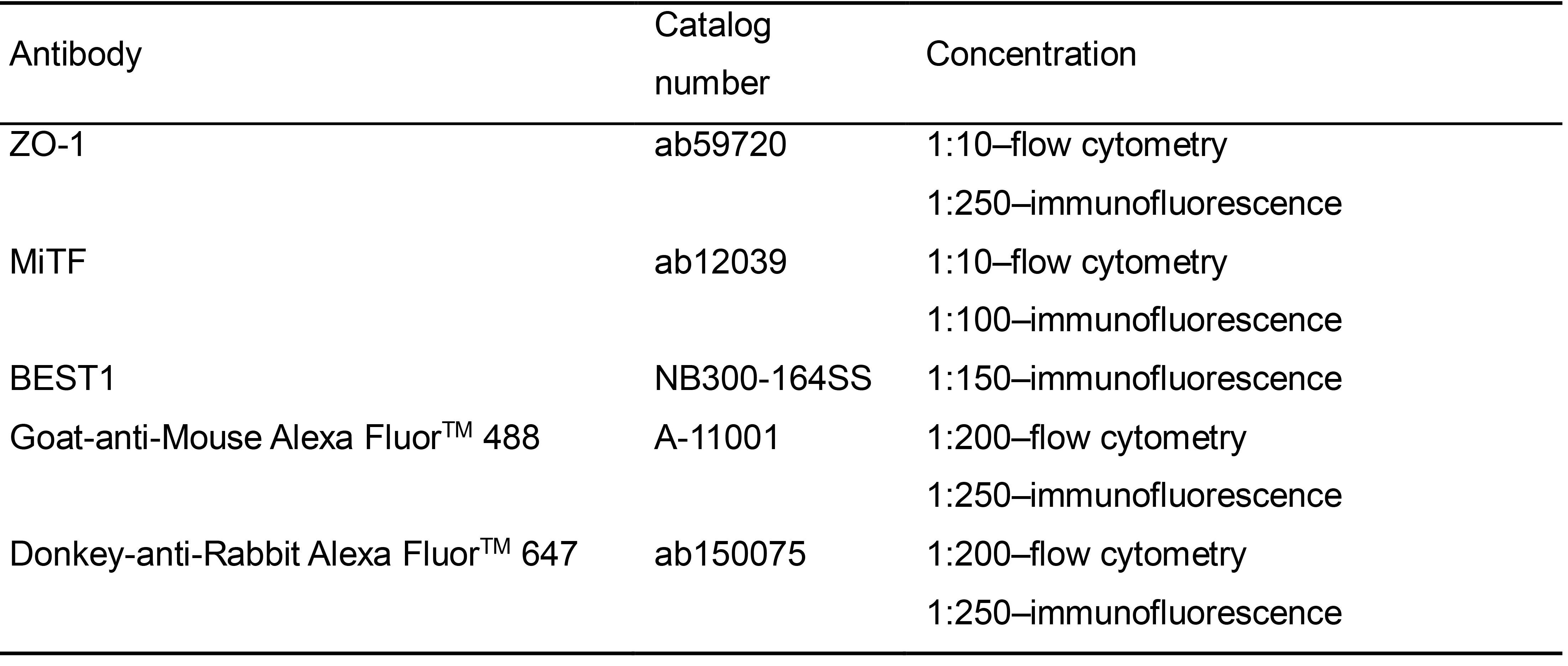
Acknowledgments
This work was supported in part by a CIRM grant GC1R-06673-B and NIH grants HG008118, HL107442, DK105541, DK112155, and EY021237. This protocol was adapted from previous work (Smith, D'Antonio-Chronowska et al., 2019).
Competing interests
Authors declare no competing interests.
Ethics
iPSC lines generated from individuals of different ethnicities (3 European Americans, 2 East Asian Americans, and 1 African American) were obtained from iPSCORE (Panopoulos et al., 2017). Donors were all females ranging from 21 to 62 years of age at the time of donation. The recruitment of these individuals was approved by the Institutional Review Boards of the University of California, San Diego, and The Salk Institute (project no. 110776ZF).
References
- Al-Zamil, W. M. and Yassin, S. A. (2017). Recent developments in age-related macular degeneration: a review. Clin Interv Aging 12: 1313-1330.
- Aoki, H., Hara, A., Nakagawa, S., Motohashi, T., Hirano, M., Takahashi, Y. and Kunisada, T. (2006). Embryonic stem cells that differentiate into RPE cell precursors in vitro develop into RPE cell monolayers in vivo. Exp Eye Res 82(2): 265-274.
- Buchholz, D. E., Hikita, S. T., Rowland, T. J., Friedrich, A. M., Hinman, C. R., Johnson, L. V. and Clegg, D. O. (2009). Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells 27(10): 2427-2434.
- Hazim, R. A., Karumbayaram, S., Jiang, M., Dimashkie, A., Lopes, V. S., Li, D., Burgess, B. L., Vijayaraj, P., Alva-Ornelas, J. A., Zack, J. A., Kohn, D. B., Gomperts, B. N., Pyle, A. D., Lowry, W. E. and Williams, D. S. (2017). Differentiation of RPE cells from integration-free iPS cells and their cell biological characterization. Stem Cell Res Ther 8(1): 217.
- Idelson, M., Alper, R., Obolensky, A., Ben-Shushan, E., Hemo, I., Yachimovich-Cohen, N., Khaner, H., Smith, Y., Wiser, O., Gropp, M., Cohen, M. A., Even-Ram, S., Berman-Zaken, Y., Matzrafi, L., Rechavi, G., Banin, E. and Reubinoff, B. (2009). Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell 5(4): 396-408.
- Klimanskaya, I., Hipp, J., Rezai, K. A., West, M., Atala, A. and Lanza, R. (2004). Derivation and comparative assessment of retinal pigment epithelium from human embryonic stem cells using transcriptomics. Cloning Stem Cells 6(3): 217-245.
- Maruotti, J., Sripathi, S. R., Bharti, K., Fuller, J., Wahlin, K. J., Ranganathan, V., Sluch, V. M., Berlinicke, C. A., Davis, J., Kim, C., Zhao, L., Wan, J., Qian, J., Corneo, B., Temple, S., Dubey, R., Olenyuk, B. Z., Bhutto, I., Lutty, G. A. and Zack, D. J. (2015). Small-molecule-directed, efficient generation of retinal pigment epithelium from human pluripotent stem cells. Proc Natl Acad Sci U S A 112(35): 10950-10955.
- Maruotti, J., Wahlin, K., Gorrell, D., Bhutto, I., Lutty, G. and Zack, D. J. (2013). A simple and scalable process for the differentiation of retinal pigment epithelium from human pluripotent stem cells. Stem Cells Transl Med 2(5): 341-354.
- Mitchell, P., Liew, G., Gopinath, B. and Wong, T. Y. (2018). Age-related macular degeneration. Lancet 392(10153): 1147-1159.
- Osakada, F., Ikeda, H., Mandai, M., Wataya, T., Watanabe, K., Yoshimura, N., Akaike, A., Sasai, Y. and Takahashi, M. (2008). Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol 26(2): 215-224.
- Osakada, F., Jin, Z. B., Hirami, Y., Ikeda, H., Danjyo, T., Watanabe, K., Sasai, Y. and Takahashi, M. (2009). In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci 122(Pt 17): 3169-3179.
- Panopoulos, A. D., D'Antonio, M., Benaglio, P., Williams, R., Hashem, S. I., Schuldt, B. M., DeBoever, C., Arias, A. D., Garcia, M., Nelson, B. C., Harismendy, O., Jakubosky, D. A., Donovan, M. K. R., Greenwald, W. W., Farnam, K., Cook, M., Borja, V., Miller, C. A., Grinstein, J. D., Drees, F., Okubo, J., Diffenderfer, K. E., Hishida, Y., Modesto, V., Dargitz, C. T., Feiring, R., Zhao, C., Aguirre, A., McGarry, T. J., Matsui, H., Li, H., Reyna, J., Rao, F., O'Connor, D. T., Yeo, G. W., Evans, S. M., Chi, N. C., Jepsen, K., Nariai, N., Muller, F. J., Goldstein, L. S. B., Izpisua Belmonte, J. C., Adler, E., Loring, J. F., Berggren, W. T., D'Antonio-Chronowska, A., Smith, E. N. and Frazer, K. A. (2017). iPSCORE: A resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports 8(4): 1086-1100.
- Pennington, K. L. and DeAngelis, M. M. (2016). Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors. Eye Vis (Lond) 3: 34.
- Reichman, S., Terray, A., Slembrouck, A., Nanteau, C., Orieux, G., Habeler, W., Nandrot, E. F., Sahel, J. A., Monville, C. and Goureau, O. (2014). From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci U S A 111(23): 8518-8523.
- Smith, E. N., D'Antonio-Chronowska, A., Greenwald, W. W., Borja, V., Aguiar, L. R., Pogue, R., Matsui, H., Benaglio, P., Borooah, S., D'Antonio, M., Ayyagari, R. and Frazer, K. A. (2019). Human iPSC-derived retinal pigment epithelium: a model system for prioritizing and functionally characterizing causal variants at AMD risk loci. Stem Cell Reports 12(6): 1342-1353.
- Sparrow, J. R., Hicks, D. and Hamel, C. P. (2010). The retinal pigment epithelium in health and disease. Curr Mol Med 10(9): 802-823.
- Vugler, A., Carr, A. J., Lawrence, J., Chen, L. L., Burrell, K., Wright, A., Lundh, P., Semo, M., Ahmado, A., Gias, C., da Cruz, L., Moore, H., Andrews, P., Walsh, J. and Coffey, P. (2008). Elucidating the phenomenon of HESC-derived RPE: anatomy of cell genesis, expansion and retinal transplantation. Exp Neurol 214(2): 347-361.
- Wong, W. L., Su, X., Li, X., Cheung, C. M., Klein, R., Cheng, C. Y. and Wong, T. Y. (2014). Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2(2): e106-116.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
D’Antonio-Chronowska, A., D’Antonio, M. and Frazer, K. A. (2019). In vitro Differentiation of Human iPSC-derived Retinal Pigment Epithelium Cells (iPSC-RPE). Bio-protocol 9(24): e3469. DOI: 10.21769/BioProtoc.3469.
Category
Stem Cell > Pluripotent stem cell > Cell differentiation
Cell Biology > Cell isolation and culture > Cell differentiation
Developmental Biology > Cell growth and fate > Differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










