- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Intracellular cAMP Measurements in Candida albicans Biofilms
Published: Vol 9, Iss 23, Dec 5, 2019 DOI: 10.21769/BioProtoc.3461 Views: 4678
Reviewed by: Alexandros AlexandratosSadri ZnaidiMichael Tscherner

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Induction and Quantification of Patulin Production in Penicillium Species
Yong Chen [...] Shiping Tian
Jun 5, 2017 9069 Views

Determination of the Cellular Ion Concentration in Saccharomyces cerevisiae Using ICP-AES
Louise Thines [...] Pierre Morsomme
Aug 20, 2020 3443 Views

A Novel and Robust Method for Investigating Fungal Biofilm
Biswambhar Biswas [...] Anil Thakur
Jan 5, 2025 1771 Views
Abstract
Candida albicans is the most common cause of fungal infections worldwide. Infection by C. albicans is closely associated with its ability to form a biofilm, closely packed communities of cells attached to the surfaces of human tissues and implanted devices, in or on the host. When tested for susceptibility to antifungals, such as polyenes, azoles, and allylamines, C. albicanscells in a biofilm are more resistant to antifungal agents than C. albicans cells in the planktonic form. Cyclic Adenosine monophosphate (cAMP) is one of the key elements for triggering hyphal and biofilm formation in C. albicans. It is hard to detect or extract molecular markers (e.g., cAMP) from C. albicans biofilms because the biofilms have a complex three-dimensional architecture with an extracellular matrix surrounding the cell walls of the cells in the biofilm. Here, we present an improved protocol that can effectively measure the level of intracellular cAMP in C. albicans biofilms.
Keywords: Candida albicansBackground
Candida albicans is a member of the fungal community in the human gastrointestinal tract, mouth, and genital tract. C. albicans does not only cause superficial diseases, but also systemic infections in immunocompromised patients (Brown et al., 2012; Kim, 2016; Lohse et al., 2018). In clinical infections of C. albicans, the fungal cells commonly form biofilms; C. albicans biofilms are associated with chronic infection and antifungal resistance (Rajendran et al., 2016). The cyclic adenosine monophosphate (cAMP) signaling pathway has been considered to be one of the most important signal transduction pathways that can cause a switch from budding to hyphal morphogenesis, a switch to hyphal growth promotes biofilm formation and invasive growth in C. albicans (Wang, 2013; Tao et al., 2017). Activation of adenylyl cyclase to produce cAMP is thought to promote hyphal induction, and maximal levels of cAMP coincide with maximal germ tube formation (Parrino et al., 2017). In brief, it is generally accepted that biofilm formation of C. albicans requires elevated levels of cAMP. Therefore, measurement of cAMP levels in C. albicans biofilms is important to understand biofilm formation and antifungal drug resistance and has the potential to be an important reference value for future drug development efforts.
C. albicans biofilms are comprised of complex, surface-associated cell populations embedded in an extracellular matrix (Lohse et al., 2018). The three-dimensional hyphal structure of biofilms and the matrix surrounding the cells serve as physical barriers that make it difficult to detect or extract specific molecular markers such as cAMP from C. albicans biofilms. Yun et al. (1998) presented a method of to measure the intracellular cAMP level of planktonic Saccharomyces cerevisiae; however, no protocol has been reported on how to measure the level of intracellular cAMP in C. albicans biofilms. This article provides an improved protocol to measure the intracellular cAMP level of C. albicans biofilms. In this protocol, the levels of intracellular cAMP of cells within biofilms are examined by using an enzyme-linked immunosorbent assay (ELISA) with modifications; this is a modification of the method originally described in Yun et al. (1998).
Materials and Reagents
- Stainless steel tweezer (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: DS0399-0001)
- Pipettes (Eppendorf, P1000, catalog number: 3120000062; P200, catalog number: 3120000054; P10, catalog number: 3120000020; P2.5, catalog number: 3120000011, respectively)
- Flasks (Duran, 50 ml, catalog numbers: 21-216-17; 100 ml, catalog number: 21-216-24, respectively)
- Cell culture dish (Thermo Fisher Scientific, Thermo ScientificTM, NuncTM EasYDishTM, catalog number: 150466)
- 1.5 ml microtubes (Axygen®, catalog number: MCT-150-C)
- Tubes (Eppendorf, 15 ml, catalog number: 0030122151; 50 ml, catalog number: 0030122178, respectively)
- Cell scraper (Costar, catalog number: 3010)
- Pipette tips (Eppendorf, 1,000 µl, catalog number: 0030000919; 200 µl, catalog number: 0030000870; 10 µl, catalog number: 0030000811, respectively)
- Aluminum foil
- Parafilm
- Strain (Candida albicans, SC5314) (ATCC, catalog number: 10231)
- Yeast extract (BD Biosciences, catalog number: 212750)
- Peptone (BD Biosciences, catalog number: 211677)
- Dextrose (BD Biosciences, catalog number: 215530)
- Agar (BD Biosciences, catalog number: 214040)
- RPMI 1640 medium (Sigma-Aldrich, catalog number: R6504)
- Sodium phosphate monobasic monohydrate (Thermo Fisher Scientific, catalog number: S369-500)
- Sodium phosphate dibasic anhydrous (Thermo Fisher Scientific, catalog number: BP332-500)
- Sodium chloride (Thermo Fisher Scientific, catalog number: S671-3)
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 59223C)
- Distilled water
- 2-(N-Morpholino)ethanesulfonic acid (MES) (Sigma-Aldrich, catalog number: M5287)
- Trichloroacetic acid (Sigma-Aldrich, catalog number: T9159)
- Ice
- Liquid nitrogen
- YPD medium
- Ether (Sinopharm Chemical Reagent Co., Ltd, catalog number: 10009318)
- cAMP Enzyme Immunoassay Kit (Sigma-Aldrich, catalog number: CA200)
- Goat Anti-Rabbit IgG Coated 96-well
- cAMP-Alkaline Phosphatase Conjugate
- cAMP EIA Antibody Rabbit Anti-cAMP
- 0.1 M Hydrochloric Acid
- 0.1 M HCI in water
- Neutralizing Reagent
- Wash Buffer Concentrate
- Cyclic AMP Standard
- p-Nitrophenyl Phosphate Substrate Solution
- Stop Solution
- Triethylamine
- Acetic Anhydride
- Plate Sealer
- MES Buffer (see Recipes)
- Sabouraud dextrose agar medium (SDA) (see Recipes)
- YPD broth (see Recipes)
- YPD agar medium (see Recipes)
- 1x PBS (see Recipes)
Equipment
- Ultrasonic cell crusher (Celton, model number: UH-500B)
- 30 °C shaking incubator (Infors, model number: Multitron Standard)
- CO2 incubator (ESCO, CelMate®, model number: CLM-170B-8-NF)
- Multiwell plate shaker (Thermo Fisher Scientific, Thermo ScientificTM, SorvallTM ST 40R, catalog number: 75004524)
- Multi-mode Microplate Reader (Molecular Devices, SpectraMax M5)
- Autoclave
- Centrifuge
Procedure
- Growth of Candida albicans and biofilm formation
- Subculture Candida albicans on Sabouraud dextrose agar medium at 35 °C.
- Pick a single colony and inoculate 10 ml of YPD medium at 30 °C in a shaking incubator (200 rpm). Grow overnight.
- Centrifuge the culture (2,100 x g, 10 min) and discard the supernatant. Use 10 ml of 1x PBS to wash the cells, centrifuge the cells for 5 min at 2,100 x g and discard the supernatant, repeat twice. Then, discard the supernatant and harvest the cells.
- Re-suspend the cells and dilute them to an optical density at 600 nm (OD600nm) of 0.4 in RPMI 1640 medium (10 ml RPMI 1640 medium for per cell culture dish).
- Inoculate the suspensions into the cell culture dishes at 37 °C without shaking (inoculum should spread to cover the entire bottom of the dish). After 2 h, gently remove the inoculum (avoid scratching away the newly forming biofilms), wash with 5 ml 1x PBS twice to remove non-adherent cells. Add 10 ml fresh RPMI 1640 medium to each dish. Then, incubate at 37 °C without shaking to form biofilms.
- Process biofilm cell samples
- Remove the medium from the dishes and gently wash the biofilms with 2 ml distilled water (slowly add distilled water using a pipet to avoid disturbing the biofilms). Add 1 ml distilled water to the dishes and scrape the dishes to isolate the biofilms, and add the samples to the 15 ml tubes. Then repeat this process, add 1 ml distilled water to collect remaining samples.
- Centrifuge the samples for 5 min at 2,100 x g at room temperature and discard the supernatant. Add 1 ml MES buffer and transfer the samples to the 1.5 ml microtubes, centrifuge the samples for 5 min at 2,100 x g, discard the supernatant.
- Then, dry the samples in a desiccator, add 10% trichloroacetic acid (not more than half volume of MES buffer), here we add 0.5 ml.
- Freeze the samples in liquid nitrogen, then thaw the samples on ice, repeat this process twice. Use an ultrasonic cell crusher to sonicate the samples under chilled conditions (twice at 130 W for 2 min).
- Add 1 ml water-saturated ether, centrifuge the samples for 5 min at 600 x g at room temperature and discard the supernatant. Repeat four times to make sure the trichloroacetic acid is extracted.
- Measure the intracellular cAMP content (Table 1)
Note: The cAMP content was measured with a cAMP enzyme immunoassay system following the manufacturer’s instructions. Allow all reagents to warm to room temperature for at least 30 min before opening and don’t use polystyrene tubes.
Table 1. The procedure for measurement of intracellular cAMP content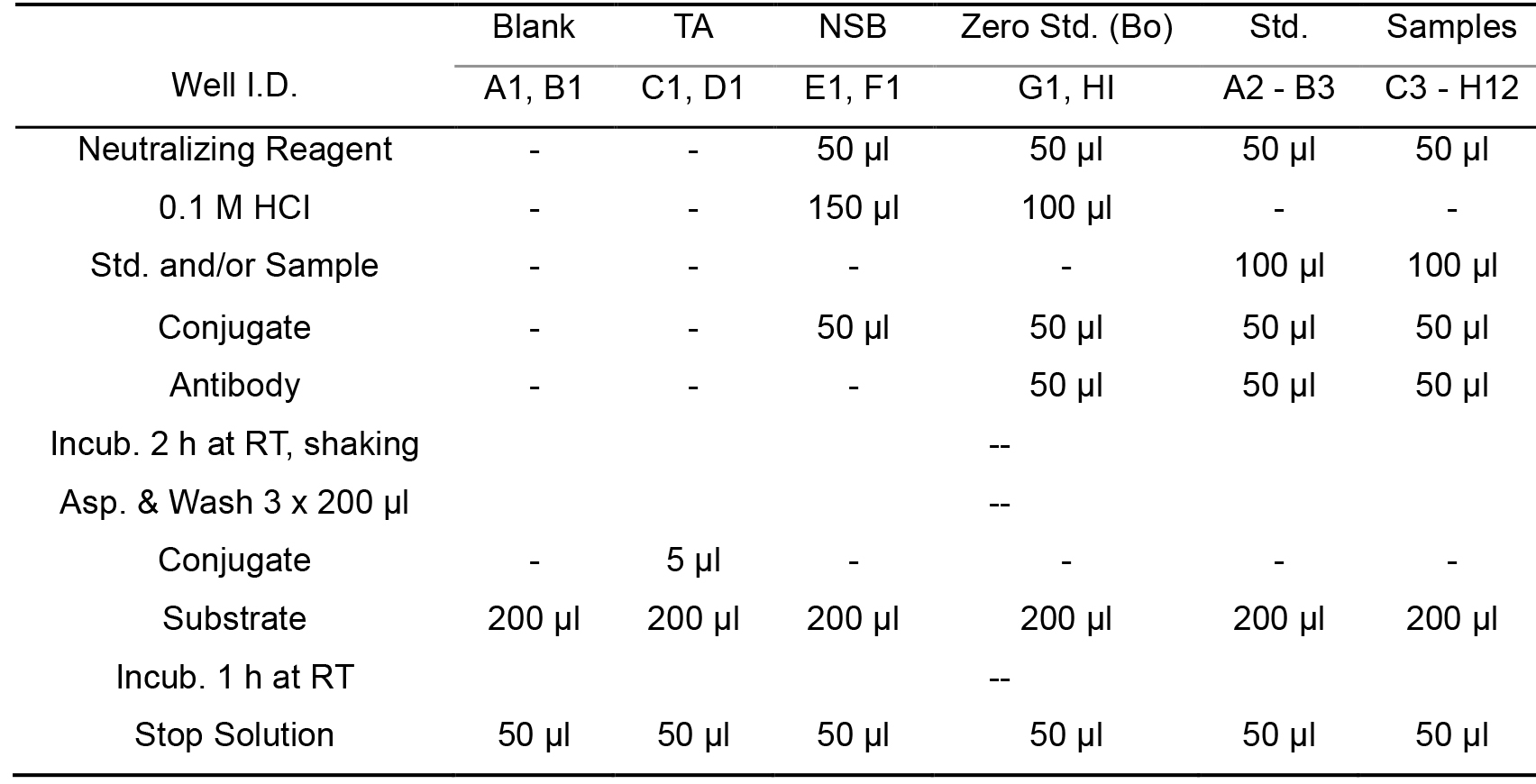
- Add 0.5 ml of acetic anhydride to 1 ml of triethylamine to prepare the Acetylation Reagent. Dilute 10 ml Wash Buffer Concentrate with 90 ml distilled water to prepare 1x Wash Buffer.
- Prepare cAMP Standard solutions:
- Label five 12 x 75 mm tubes from 1 to 5.
- Add 990 µl of 1 M HCI into tube 1 and 750 µl into tubes 2-5. Add 10 µl of the Cyclic AMP Standard to tube 1 and mix thoroughly.
- Then, transfer 250 µl solution from Tube 1 to Tube 2. Dilute the cAMP from Tube 2 to Tube 5 in series. The concentration of cAMP in Tubes 1 through 5 will be 20, 5, 1.25, 0.312, and 0.078 pmol/ml, respectively (Table 2).
- Label one 12 x 75 mm tube as the Zero Standard/NSB tube.
Table 2. Preparation of cAMP standard solution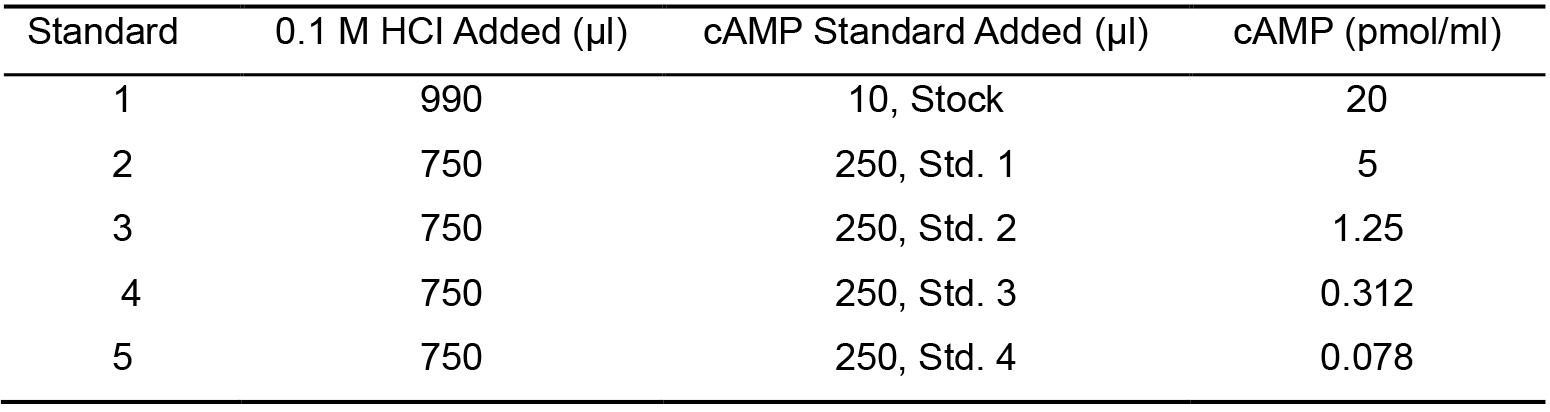
- Add 50 µl Neutralizing Reagent into each well of the ELISA plate, except wells A1-D1 (Figure 1). Add 100 µl 0.1 M HCI into the wells E1-H1, respectively. Add 100 µl of standard solutions 1-5 into wells A2-B3.
- Add 100 µl of the samples into the appropriate wells.
- Add 50 µl 0.1 M HCI into wells E1-F1 (NSB wells, non specific binding wells).
- Add 50 µl blue cAMP-Alkaline Phosphatase Conjugate into each well except wells A1-D1. Add 50 µl yellow cAMP EIA antibody into all of the wells except A1-F1 (Figure 1).
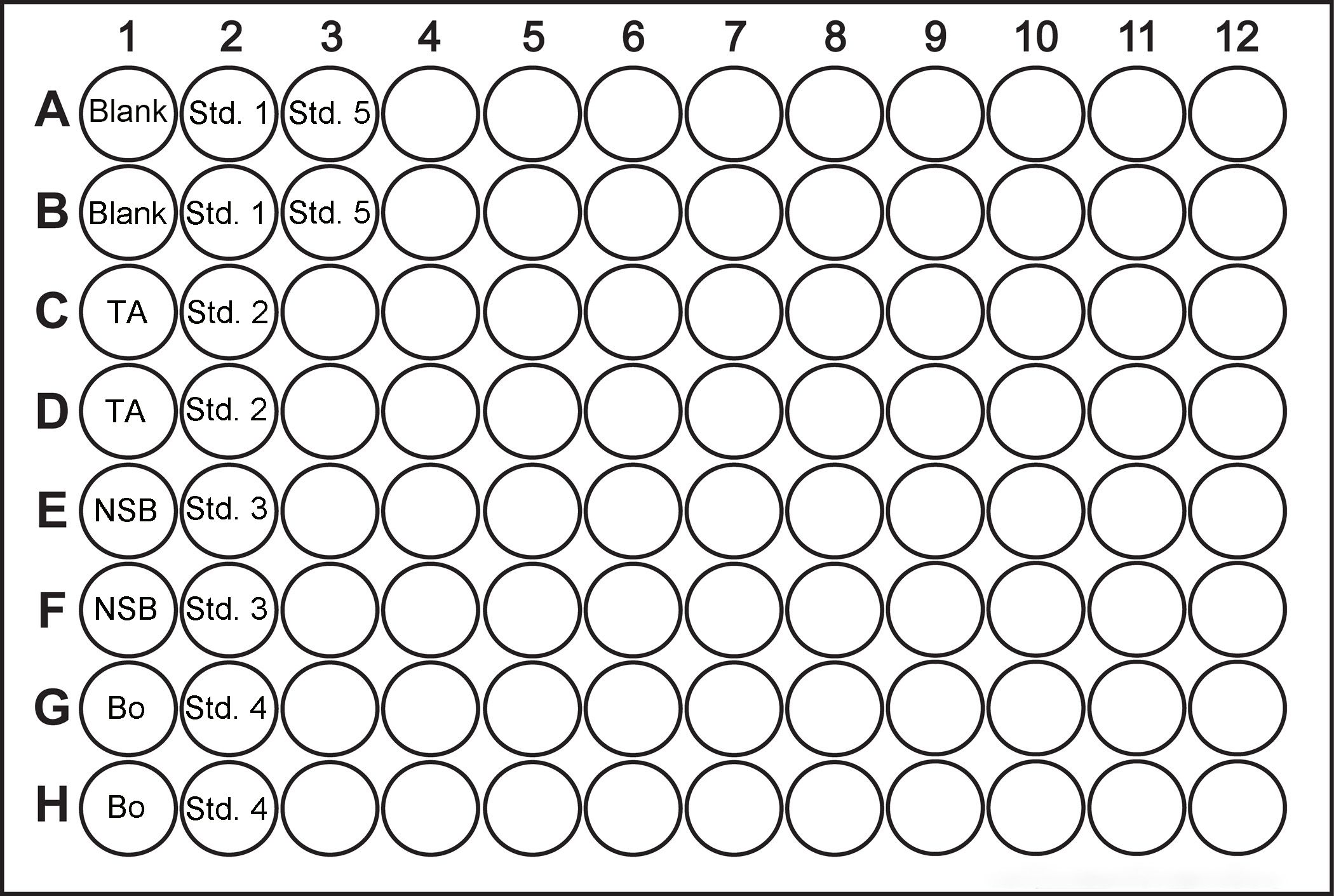
Figure 1. Plate layout for the cAMP measurement - Incubate the plate at RT for 2 h on a plate shaker (500 rpm).
- Empty the contents of the wells and use 1x Wash Buffer to wash every well. Repeat twice and discard the remaining wash buffer.
- Add 50 µl of blue cAMP-Alkaline Phosphatase Conjugate to the TA wells. Add 200 µl of the p-Nitrophenyl Phosphate Substrate Solution to every well.
- Incubate the plate at RT for 1 h without shaking.
- Add 50 µl of Stop Solution to every well. Read the optical density (OD) at 405 nm, preferably with correction between 570 and 590 nm.
Data analysis
Calculate the concentration of cAMP as follows (Table 3):
- Calculate the Average Net Optical Density (OD) bound for each standard and sample:
Average Net OD = Average Bound OD – Average NSB OD - Calculate the binding of each pair of standard wells as a percentage of the maximum binding wells (Bo):
Percent Bound = Net OD/Net Bo OD x 100 - Use the Logit-Log paper plot Percent Bound versus Concentration of cAMP for the standards and draw a straight line through those points. Then, determine the concentration of cAMP by interpolation.
Table 3. Typical results for calculating the concentration of cAMP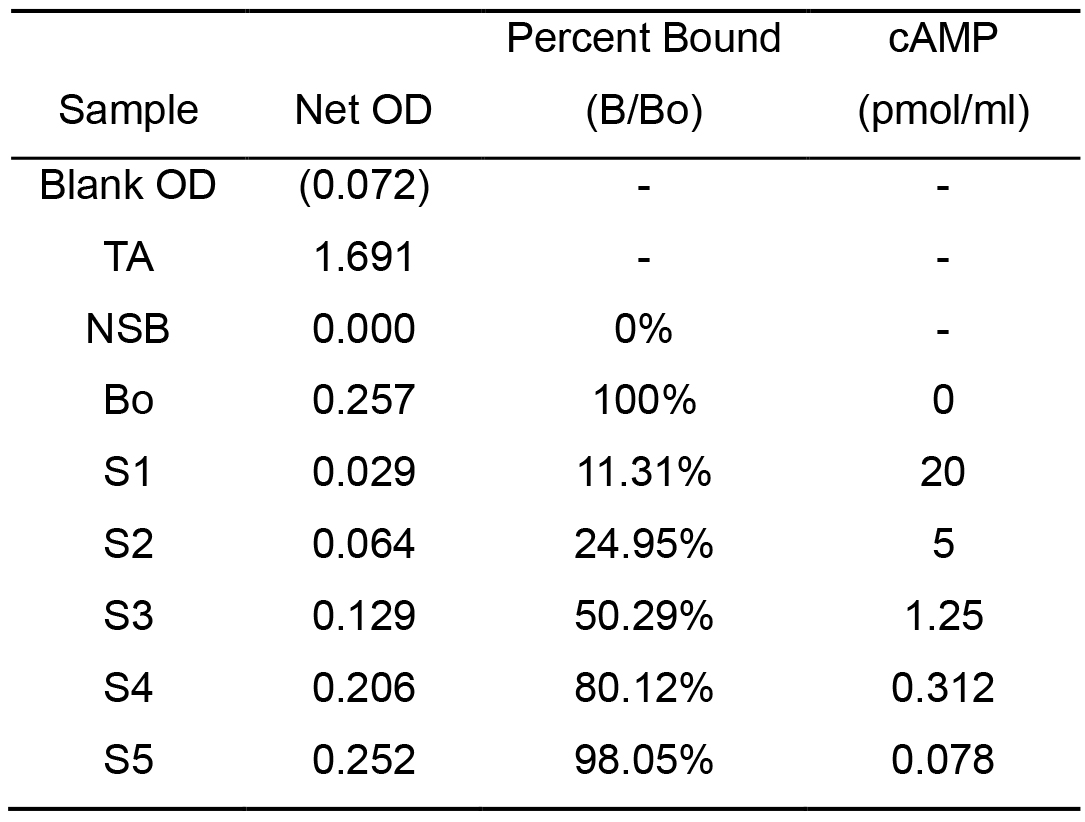
Recipes
- MES Buffer (1 L)
21.3 g MES
5 M NaOH (adjust pH to 6.0)
1 L Distilled water
Autoclave (120 °C, 120 kPa for 20 min) - SDA agar medium (1 L)
10 g Peptone
40 g Dextrose
20 g Agar (2%)
1 L Distilled water
Autoclave (120 °C, 120 kPa for 20 min) - YPD broth (1 L)
10 g Yeast extract (1%)
20 g Peptone (2%)
20 g Dextrose (2%)
1 L Distilled water
Autoclave (120 °C, 120 kPa for 20 min) - YPD agar medium (1 L)
10 g Yeast extract (1%)
20 g Peptone (2%)
20 g Dextrose (2%)
20 g Agar (2%)
1 L Distilled water
Autoclave (120 °C, 120 kPa for 20 min) - 1x PBS
2.62 g Sodium phosphate monobasic monohydrate
11.5 g Sodium phosphate dibasic anhydrous
43.84 g Sodium chloride
500 ml Distilled water
Adjust pH to 7.2 (1 M NaOH)
Autoclave (120 °C, 120 kPa for 20 min)
Acknowledgments
This protocol was adapted from our previously published work (Chen et al., 2018). This work was supported by the National Natural Sciences Foundation of China under Grant No. 81970945 and No. 81371156 and the Foundation of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2018-87).
Competing interests
The authors declare that there are no conflicts of interest or competing interests.
References
- Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G. and White, T. C. (2012). Hidden killers: human fungal infections. Sci Transl Med 4(165): 165rv13.
- Kim, J. Y. (2016). Human fungal pathogens: Why should we learn? J Microbiol 54(3): 145-148.
- Lohse, M. B., Gulati, M., Johnson, A. D. and Nobile, C. J. (2018). Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol 16(1): 19-31.
- Parrino, S. M., Si, H., Naseem, S., Groudan, K., Gardin, J. and Konopka, J. B. (2017). cAMP-independent signal pathways stimulate hyphal morphogenesis in Candida albicans. Mol Microbiol 103(5): 764-779.
- Rajendran, R., Sherry, L., Nile, C. J., Sherriff, A., Johnson, E. M., Hanson, M. F., Williams, C., Munro, C. A., Jones, B. J. and Ramage, G. (2016). Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin Microbiol Infect 22(1): 87-93.
- Tao, L., Zhang, Y., Fan, S., Nobile, C. J., Guan, G. and Huang, G. (2017). Integration of the tricarboxylic acid (TCA) cycle with cAMP signaling and Sfl2 pathways in the regulation of CO2 sensing and hyphal development in Candida albicans. PLoS Genet 13(8): e1006949.
- Wang, Y. (2013). Fungal adenylyl cyclase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog 9(10): e1003612.
- Yun, C. W., Tamaki, H., Nakayama, R., Yamamoto, K. and Kumagai, H. (1998). Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 252(1): 29-33.
- Chen, S., Xia, J., Li, C., Zuo, L., Wei, X. (2018). The possible molecular mechanisms of farnesol on the antifungal resistance of C. albicans biofilms: the regulation of CYR1 and PDE2. BMC Microbiol 4;18(1): 203.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jiang, L., Chen, S., Sun, K., Zhou, P. and Wei, X. (2019). Intracellular cAMP Measurements in Candida albicans Biofilms. Bio-protocol 9(23): e3461. DOI: 10.21769/BioProtoc.3461.
Category
Microbiology > Microbial biochemistry > Other compound
Microbiology > Microbial biofilm > Biofilm culture
Biochemistry > Other compound > cAMP
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









