- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Phospho-protein Analysis in Adherent Cells Using Flow Cytometry
Published: Vol 9, Iss 20, Oct 20, 2019 DOI: 10.21769/BioProtoc.3395 Views: 5999
Reviewed by: Ralph Thomas BoettcherRAVI THAKURMasashi Asai

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Automated Layer Analysis (ALAn): An Image Analysis Tool for the Unbiased Characterization of Mammalian Epithelial Architecture in Culture
Christian Cammarota [...] Tara M. Finegan
Apr 20, 2024 4223 Views

Quantitative Measurement of the Kinase Activity of Wildtype ALPK1 and Disease-Causing ALPK1 Mutants Using Cell-Free Radiometric Phosphorylation Assays
Tom Snelling
Nov 20, 2024 1581 Views

Accurate Identification of Cell Cycle Stages in RPE1 Cells Using the ImmunoCellCycle-ID Method
Syon Reddy [...] Aussie Suzuki
Aug 5, 2025 1870 Views
Abstract
Protein phosphorylation is one of the most important post-translational modifications, which acts as a reversible on or off switch for the activity of a large number of proteins. Analyzing the phosphorylation status of different proteins can reveal the alterations in the state of the cells in response to cellular damage, cancer and pharmaceutical drugs. Techniques such as mass spectrometry, radiolabeling, 2D-gel electrophoresis and western blotting are used to quantify protein phosphorylation. These assays can quantify phosphorylation in the bulk population of cells, however, flow cytometry can couple cell surface marker expression data with phosphorylation data to understand differential signaling in a sub-population within a heterogeneous population of cells. Our protocol describes the use of flow-cytometry for rapid and single cell-based quantification of intracellular phospho-protein with the help of anti-phospho protein specific antibody.
Keywords: Phospho-proteinBackground
Protein phosphorylation is one of the most intensively studied post translation modifications. Protein phosphorylation acts as an “on” or “off” switch for the target protein activity and thereby modulates a large number of pathways and biological processes (Hunter, 1995). Phosphorylation of proteins is reversibly mediated by the action of two classes of enzymes namely; protein kinases and protein phosphatases, which together constitute the largest enzyme family, spanning 2% of the human genome (Venter, 2001; Manning et al., 2002; Alonso et al., 2004). It is estimated that one in three proteins undergoes phosphorylation during its lifetime. Given the scale of impact and importance of protein phosphorylation in dictating protein function, tight regulation of phosphorylation by protein kinases and phosphatases play vital role in signal transduction, cell differentiation, development, cell cycle control and metabolism (Delom and Chevet, 2006; Ardito et al., 2017). Protein kinases can be divided into two major groups based on the site of phosphorylation; serine/threonine kinases and tyrosine kinases (Roskoski, 2015). Whereas up to 86.4% of all phosphorylation modifications occur at serine residues, followed by 11.8% at threonine residues, less than 2% occur at tyrosine residues (Ardito et al., 2017).
The analysis of phospho-proteins is coupled with a myriad number of complications. Majority of proteins exist as a heterogeneous population, wherein the phosphorylated fraction is generally low in abundance and it exists in several different phosphorylated forms. The reversible nature of phosphorylation due to activity of phosphatases further necessitates crucial precautions during the processing of samples (Delom and Chevet, 2006). Mass spectroscopy analysis enables identification and quantification of post-translation modifications of proteins at a large scale (Steen et al., 2006; Junger and Aebersold, 2014; Pan et al., 2015). Two-dimensional gel electrophoresis can exploit the difference in isoelectric point of phosphorylated variant of a protein to quantify phosphorylated fraction (Guy et al., 1994). Use of phospho-protein specific antibodies conjugated with fluorophores allows single cell based rapid quantification with the help of flow cytometry (Krutzik and Nolan, 2003). Flow cytometry offers reduction in the number of processing steps as compared to western blotting and enables multiplex analysis of different types of cells and phospho-proteins (Krutzik et al., 2004; Davies et al., 2016). Whereas traditional biochemical assays can only quantify phosphorylation in the bulk population of cells but cannot determine the signaling differences in a sub-population within a heterogeneous population of cells, flow cytometry can couple cell surface marker expression data with phosphorylation data to understand differential response of different cell types to a ligand or inhibitor (Krutzik et al., 2004). Phospho-protein flow cytometry can be used to study signaling pathways in stem cells (Sonowal et al., 2013), cancer cells (Kumar et al., 2017) and understand cell-cell interaction (Kumar et al., 2018). We developed a reliable method to analyze phospho-proteins in adherent cells. Adherent cells require an additional step, where the cells have to be enzymatically detached from the cell growth surface and can lead to inactivation of phospho-proteins. This protocol described here can be used to successfully quantify the intracellular phospho-proteins in adherent cell types.
Materials and Reagents
- Bio-hazard waste container (Tarsons, catalog number: 583254)
- Cell culture dish, 100 x 20 mm (Eppendorf, catalog number: 0030702115)
- T25 flasks (Eppendorf, catalog number: 0030710118)
- FACS tubes (Corning, catalog number: 352063)
- Graduated centrifuge tubes, 15 ml (Tarsons, catalog number: 546021)
- Graduated centrifuge tubes, 50 ml (Tarsons, catalog number: 546041)
- Graduated 0.2-10 μl micro-tips (Tarsons, catalog number: 521000)
- Graduated 2-200 μl micro-tips (Tarsons, catalog number: 521010)
- Graduated 200-1000 μl micro-tips (Tarsons, catalog number: 521020)
- Serological glass pipettes, 10 ml (Himedia, catalog number: CG316-1x10NO)
- Sterile filtration unit (0.22 μm) (Thermo Fisher Scientific, catalog number: 450-0020)
- Storage vial, 5 ml (Tarsons, catalog number: 523070)
- MDA-MB-231 breast cancer cell line (NCCS, Pune)
- 10x Trypsin (2.5%) (Thermo Fisher Scientific, catalog number: 15090-046)
- Anti-Smad1 (pS463/pS465)/Smad8 (pS465/pS467) PE conjugated antibody (BD Biosciences, catalog number: 562509)
- Bovine serum albumin (BSA) (Himedia, catalog number: MB083-5G)
- De-ionized water (dH2O) (Merck, Elix Type 2 pure water)
- Di-sodium hydrogen phosphate (Na2HPO4) (Merck, catalog number: 61755005001046)
- DMEM-high glucose with L-Glutamine medium (DMEM-HG) (Sigma-Aldrich, catalog number: D5648-1L)
- Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, catalog number: 10270)
- Formaldehyde solution 37-41% (w/v) (Merck, catalog number: 61780805001730)
- Hydrochloric acid 35% (HCl) (Merck, catalog number: 61762505001730)
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418-250ML)
- LDN193189 hydrochloride (Sigma-Aldrich, catalog number: SML0559-5MG)
- Methanol (Merck, catalog number: 82228305031730)
- Potassium chloride (KCl) (Merck, catalog number: 61779205001730)
- Potassium dihydrogen phosphate (KH2PO4) (Merck, catalog number: 60487305001730)
- Sodium bicarbonate (NaHCO3) (Sigma-Aldrich, catalog number: S5761)
- Sodium chloride (NaCl) (Merck, catalog number: 1.93206.0521)
- Sodium hydroxide (NaOH) (Merck, catalog number: 61843805001730)
- Trypan blue (Sigma-Aldrich, catalog number: T6146)
- 100x Penicillin (10,000 Units/ml)-Streptomycin (10,000 Units/ml) antibiotic (Thermo Fisher Scientific, catalog number: 15140-122)
- DMEM medium (see Recipes)
- PBS (see Recipes)
- 1x Trypsin (see Recipes)
- Heat inactivated FBS (see Recipes)
- 0.4% trypan blue (see Recipes)
- 4% formaldehyde solution (see Recipes)
- Staining buffer (see Recipes)
- LDN193189 Hydrochloride stock (see Recipes)
Equipment
- Hemocytometer (Sigma-Aldrich, catalog number: Z359629-1EA)
- Analytical balance (Sartorius, Quintix Analytical Balance 60, 120 g x 0.01, 0.1 mg)
- Autoclave*
- Biosafety cabinet class II (Thermo Fisher Scientific, model: 1300 series A2)
- Centrifuge (Thermo Fisher Scientific, Sorvall Legend X1R centrifuge, catalog number: 75004260)
- CO2 incubator (Thermo Fisher Scientific, Hera Cell 150i)
- Flow cytometer (Becton Dickinson, FACS calibur and BD cell quest software)
- Inverted microscope with camera (Zeiss, Axio Vert A1)
- Vortex mixer (IKA, model: MS-3D)
- Micropipettes (Thermo Fisher Scientific, Finnpipette F2–10 μl, 100 μl and 1,000 μl)
- Pipette controller (Socorex, Profiller 446)
- Magnetic Stirrer (Tarsons, catalog number: 6090)
- 4 °C refrigerator*
- -20 °C refrigerator*
*Note: These items can be ordered from any qualified company. - -80 °C refrigerator (Thermo Fisher Scientific, FORMA 88000 Series)
- Water bath (Thermo Fisher Scientific, Labline water bath)
Software
- FlowJo Software (FlowJo, LLC)
Procedure
For this study, MDA-MB-231 breast cancer cell line was purchased from National Centre for Cell Science (NCCS, Pune, India). The cells were cultured in cell growth medium (Recipe 1) in a CO2 incubator maintained at 5% CO2 and 37 °C.
Note: Purchase cells from a recognized cell center and maintain them according to the instructions provided by the supplier.
- Maintenance of cells
Note: The following steps are to be performed inside a biosafety cabinet class II following aseptic culture techniques.
Once the cells reach a confluency of 80%-90%, subculture them as follows:- Aspirate the spent medium and collect it in a sterile 15 ml centrifuge tube.
- Wash twice with 1x PBS (Recipe 2).
- Add 500 μl of 1x Trypsin (Recipe 3) to the cells and incubate at 37 °C for 2-3 min.
Notes:
- Keep trypsin at room temperature for 5-10 min after retrieving from 4 °C before use.
- The amount of trypsin recommended is for 60 mm dish or T25 flasks. For larger surface area, adjust the volume of trypsin used accordingly.
- Gently tap along the side of dish to dislodge the cells. Do not leave cells in trypsin for more than 5 min.
- Add 1-2 ml spent medium to neutralize the effect of trypsin and collect the cells in a labeled 15 ml centrifuge tube.
- Centrifuge the cells at 300 x g for 5 min at 4 °C to obtain the cell pellet. Discard the supernatant and re-suspend the pellet in 1 ml of fresh growth medium.
- Resuspend cells properly and take out small volume of cells (50 μl) for counting.
- Add an equal volume of 0.4% trypan blue (Recipe 5), mix and count viable cells using hemocytometer.
- Seed cells in a 100 mm dish at a density of 5,000 cells/cm2 in 10 ml of growth medium. Allow the cells to attach overnight.
Note: This seeding density is recommended for MDA-MB-231 cell line. Depending on the growth rate of cells used, seed cells at a density that will give at least 1 x 106 cells at the time of harvesting the cells for experiment. - After 24 h, check the cells under the microscope for any contamination, add the stimulator/drugs/molecules for treatment at required concentration, and incubate the cells for the required amount of time in a CO2 incubator at 37 °C.
Note: For the current analysis, the cells were treated with 1 μM LDN193189 hypochloride (Recipe 8) for 24 h.
- Harvesting of cells
Notes:- Before starting the experiment go through the entire protocol and ensure the availability of all required materials and the temperature of water bath has reached 37 °C.
- Pre-warm PBS at 37 °C for 30 min and pre-warm trypsin at 37 °C for no longer than 2-3 min.
- Temperature of the cells should be maintained at 37 °C until the fixation step is completed, as fluctuation in temperature can lead to dephosphorylation of the phospho-proteins.
- Perform the following steps in a 37 °C water bath, but not in an incubator to ensure maintenance of uniform temperature during the procedure.
- Handle the dish carefully, so that the dish is not contaminated with the water from the water bath.
- Take the cells out from the CO2 incubator, discard the media immediately by pouring directly to bio-hazard waste container and place the dish in a 37 °C water bath.
- Add 10 ml of pre-warmed 1x PBS along the wall of dish and swirl the dish gently to wash off the growth medium completely.
- Discard the PBS and remove any remaining PBS by gently tapping the dish on tissue towel for 3-4 s.
- Immediately place the dish in the water bath and add 900 μl of pre-warmed 1x Trypsin (see Recipes). Gently swirl the dish to ensure uniform distribution of trypsin. Incubate for 1-3 min depending on the cells type.
Note: The amount of trypsin recommended in this step is for 100 mm dish. Adjust the volume according to the size of the dish used for experiment. - Gently tap the dish on the sides to ensure proper detachments of cells. A slight turbidity can be observed after trypsinization.
- Add 100 μl of pre-warmed FBS directly to the dish to neutralize the effect of trypsin and mix by pipetting.
- Fixation of cells
Notes:- Perform Steps C2-C4 at room temperature.
- Some phospho-proteins and/or antibodies might require a different fixation buffer, the user should check with the manufacturer before proceeding with the fixation of cells.
- Add 1 ml 4% formaldehyde (Recipe 6) directly to the cells, mix well by pipetting 3-4 times and transfer the cell suspension into a FACS tube.
Note: 4% paraformaldehyde can also be used instead of 4% formaldehyde for fixation following the same steps as mentioned above. - Vortex the cell suspension for about 10-15 s at 2500 rpm and incubate for 10 min at room temperature.
- Add 2 ml of staining buffer (Recipe 7) to the cell suspension and centrifuge at 500 x g for 5 min at 4 °C. Discard the supernatant without disturbing the pellet.
- Wash the cells by resuspending the pellet in 2 ml of ice-cold 1x PBS. Centrifuge at 500 x g for 5 min at 4 °C and discard the supernatant.
- Permeabilization of cells
- While vortexing at a speed of 2500 rpm, add ice-cold 100% methanol (1 ml for 1 x 106 cells) drop-wise to the cell pellet.
Note: Keep 100% methanol on ice for 2 h or alternatively at -20 °C overnight prior to starting the experiment. - After addition of methanol, cover the tube with a cap to avoid methanol loss and incubate for at least 1 h at 4 °C (Figure 1).
- After methanol addition, cells can be stored for a week at -20 °C.
- Although most of the phospho-proteins and antibodies can work well with methanol mediated permeabilization, some phospho-proteins and/or antibodies might require a different permeabilization buffer, the user should check with the manufacturer before proceeding further.

Figure 1. Procedure describing sample processing for phospho protein staining
- While vortexing at a speed of 2500 rpm, add ice-cold 100% methanol (1 ml for 1 x 106 cells) drop-wise to the cell pellet.
- Phospho-protein staining and flow cytometry analysis
- Add 4 ml of staining buffer to the cells in methanol, centrifuge at 500 x g for 5 min at 4 °C and discard the supernatant gently without disturbing the cell pellet.
- Resuspend the cell pellet in 2 ml of staining buffer. Centrifuge at 500 x g for 5 min at 4 °C and discard the supernatant.
- Resuspend the pellet in 100 μl of staining buffer and distribute 50 μl of cell suspension per FACS tube for further staining.
Note: Make sure that at least 5 x 105 cells per tube are used for further staining for phospho-proteins. - Add phospho-specific primary unconjugated or fluorescent conjugated antibody to the cells according to the specifications of the manufacturer to one of the tubes and add isotype control to the other tube.
- Gently vortex the suspension for 10 s and incubate at room temperature in the dark for 1 h.
- Add 2 ml of staining buffer, centrifuge at 500 x g for 5 min at 4 °C and discard the supernatant carefully.
- If a fluorescent conjugated antibody was used in Step E4, proceed directly to Step E9. If an unconjugated primary antibody was used, resuspend the cell pellet in 50 μl of staining buffer. Add fluorescent dye conjugated secondary antibody and incubate at room temperature in the dark for 1 h.
- Add 2 ml of staining buffer, centrifuge at 500 x g for 5 min at 4 °C and discard the supernatant carefully.
- Resuspend the pellet in 300 μl of staining buffer and keep on ice.
- Analyze the samples in a flow cytometer using proper settings for the fluorescence detection channels (Figure 2).
Note: Ensure that the excitation and emission wavelength of the fluorophore conjugated-antibody is compatible with the flow cytometer used.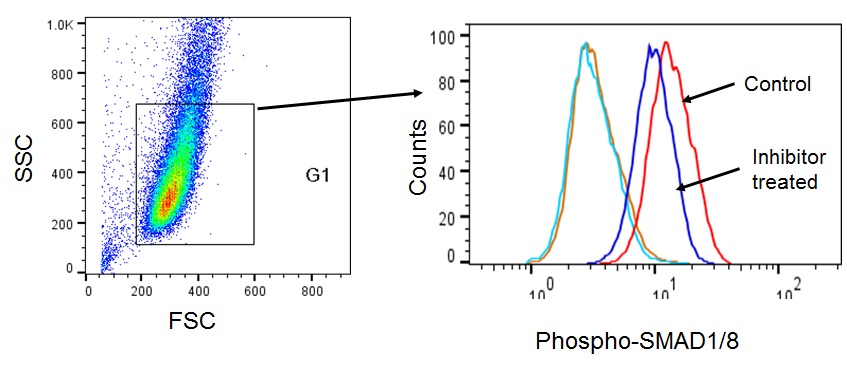
Figure 2. Flow cytometric analysis of phospho-SMAD1/8. MDA-MB-231 cells were stained with PE conjugated anti-Smad1 (pS463/pS465)/Smad8 (pS465/pS467) antibody (Phospho-SMAD1/8). Cells were left untreated (Control) or treated with BMP inhibitor LDN193189 (Inhibitor treated). Orange line is the unstained control for control cells and light blue line is the unstained control for inhibitor treated cells.
- DMEM medium
DMEM-HG (high glucose) basal medium:
10 g DMEM-HG media powder
3.7 g sodium bicarbonate
10 ml 100x Penicillin-Streptomycin antibiotic solution
Make up the volume to 1 L with autoclaved deionized H2O
Adjust pH to ~7.0 with 5 M HCl as pH tends to increase after filtration
Sterilize by filtration using a sterile filtration unit (0.22 μm)
Can be stored at 4 °C for up to 2 months
Cell growth medium:
10% FBS in DMEM-HG basal medium
Sterilize by filtration using a sterile filtration unit (0.22 μm)
Store at 4 °C and use within two weeks - Phosphate buffered saline (PBS)
10x PBS:
80 g NaCl
2 g KCl
14.4 g Na2HPO4
2.4 g KH2PO4
Add all salts in 800 ml of autoclaved deionized dH2O, and allow it to stir for 1 h at room temperature using a magnetic stirrer
Make up the volume to 1 L with deionized water
Sterilize by filtration using a sterile filtration unit (0.22 μm)
Store at room temperature
1x PBS:
Dilute 10x PBS ten times (1 ml in 10 ml) in H2O
Adjust pH to 7.4 with 5 M NaOH and autoclave at 121 °C for 20 min - 1x Trypsin (0.25%)
1 ml of 10x trypsin
9 ml of sterile PBS
Prepare 5 ml aliquots for use
Store aliquots at -20 °C for long term storage
Thaw at room temperature and store at 4 °C for short term use up to 1 week\ - Heat inactivated FBS
Heat inactivate FBS by incubating at 56 °C for 1 h in a water bath and store at -20 °C as 50 ml aliquots - 0.4% trypan blue
0.04 g of Trypan blue (dye composition 40%)
10 ml of 1x PBS
Filter using a sterile filtration unit (0.22 μm)
Store aliquots at 4 °C for long term storage - 4% formaldehyde solution
Dilute 37%-41% formaldehyde solution 10 times in 1x PBS
1 ml of 37%-41% formaldehyde solution
9 ml of 1x PBS
Mix it well by inverting 4-5 times
Prepare immediately before use
Keep at room temperature - Staining Buffer
0.5 g BSA
100 ml of 1x PBS
Stir to dissolve
Store at 4 °C up to 1 week - LDN193189 Hydrochloride stock
Dissolve 5 mg in 1.23 ml of DMSO to obtain 10 mM stock
Store at -20 °C as aliquots of 20 μl each
Thaw on ice before use - Alonso, A., Sasin, J., Bottini, N., Friedberg, I., Friedberg, I., Osterman, A., Godzik, A., Hunter, T., Dixon, J. and Mustelin, T. (2004). Protein tyrosine phosphatases in the human genome. Cell 117(6): 699-711.
- Ardito, F., Giuliani, M., Perrone, D., Troiano, G. and Muzio, L. L. (2017). The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int J Mol Med 40(2): 271-280.
- Davies, R., Vogelsang, P., Jonsson, R. and Appel, S. (2016). An optimized multiplex flow cytometry protocol for the analysis of intracellular signaling in peripheral blood mononuclear cells. J Immunol Methods 436: 58-63.
- Delom, F. and Chevet, E. (2006). Phosphoprotein analysis: from proteins to proteomes. Proteome Sci 4: 15.
- Guy, G. R., Philip, R. and Tan, Y. H. (1994). Analysis of cellular phosphoproteins by two-dimensional gel electrophoresis: applications for cell signaling in normal and cancer cells. Electrophoresis 15(3-4): 417-440.
- Hunter, T. (1995). Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80(2): 225-236.
- Junger, M. A. and Aebersold, R. (2014). Mass spectrometry-driven phosphoproteomics: patterning the systems biology mosaic. Wiley Interdiscip Rev Dev Biol 3(1): 83-112.
- Krutzik, P. O., Irish, J. M., Nolan, G. P. and Perez, O. D. (2004). Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol 110(3): 206-221.
- Krutzik, P. O. and Nolan, G. P. (2003). Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A 55(2): 61-70.
- Kumar, A., Anand, T., Bhattacharyya, J., Sharma, A. and Jaganathan, B. G. (2018). K562 chronic myeloid leukemia cells modify osteogenic differentiation and gene expression of bone marrow stromal cells. J Cell Commun Signal 12(2): 441-450.
- Kumar, A., Bhattacharyya, J. and Jaganathan, B. G. (2017). Adhesion to stromal cells mediates imatinib resistance in chronic myeloid leukemia through ERK and BMP signaling pathways. Sci Rep 7:9535.
- Manning, G., Whyte, D. B., Martinez, R., Hunter, T. and Sudarsanam, S. (2002). The protein kinase complement of the human genome. Science 298(5600): 1912-1934.
- Pan, L., Wang, L., Hsu, C. C., Zhang, J., Iliuk, A. and Tao, W. A. (2015). Sensitive measurement of total protein phosphorylation level in complex protein samples. Analyst 140(10): 3390-3396.
- Roskoski, R. (2015). A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res 100: 1-23.
- Sonowal, H., Kumar, A., Bhattacharyya, J., Gogoi, P. K. and Jaganathan, B. G. (2013). Inhibition of actin polymerization decreases osteogeneic differentiation of mesenchymal stem cells through p38 MAPK pathway. J Biomed Sci 20: 71.
- Steen, H., Jebanathirajah, J. A., Rush, J., Morrice, N. and Kirschner, M. W. (2006). Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics 5(1): 172-181.
- Venter, J. C. (2001). The sequence of the human genome (vol 292, pg 1304, 2001). Science 292(5523): 1838-1838.
Data analysis
The flow cytometry data of the phospho-protein staining was analyzed using FlowJo software. Cell population was selected by gating in FSC vs. SSC plot to remove the cell debris. Histograms were plotted to show the differences between isotype/unstained controls versus stained sample and treated versus untreated conditions. This protocol shows representative data for one phospho-protein and does not include any statistical analysis.
Notes
It is important to use antibodies that were tested for flow cytometry and detect only phospho form of the protein of interest.
Recipes
Acknowledgments
This work was supported by funds from Indian Council of Medical Research (ICMR), Govt. of India and Department of Science and Technology (DST), Govt. of India. RS and AS are supported by fellowship from Ministry of Human Resource and Development (MHRD), Govt. of India. This protocol was adapted from Kumar et al. (2017 and 2018).
Competing interests
The authors declare no competing interests.
References
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sharma, R., Sharma, A., Kumar, A. and Jaganathan, B. G. (2019). Phospho-protein Analysis in Adherent Cells Using Flow Cytometry. Bio-protocol 9(20): e3395. DOI: 10.21769/BioProtoc.3395.
Category
Cell Biology > Cell signaling > Phosphorylation
Cell Biology > Cell imaging > Fixed-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









