- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analyzing the Functionality of Non-native Hsp70 Proteins in Saccharomyces cerevisiae
Published: Vol 9, Iss 19, Oct 5, 2019 DOI: 10.21769/BioProtoc.3389 Views: 4398
Reviewed by: Juan Facundo Rodriguez AyalaAnna A. ZorinaFernando A Gonzales-Zubiate

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Imaging Gene Expression Dynamics in Pseudomonas fluorescens In5 during Interactions with the Fungus Fusarium graminearum PH-1
Rosanna C. Hennessy [...] Stefan Olsson
Jun 20, 2019 5357 Views

Efficient Generation of Genome-wide Libraries for Protein–ligand Screens Using Gibson Assembly
Tamara Sternlieb [...] Igor Cestari
Nov 20, 2022 3436 Views

Purification of Human Cytoplasmic Actins From Saccharomyces cerevisiae
Brian K. Haarer [...] Jessica L. Henty-Ridilla
Dec 5, 2023 1748 Views
Abstract
Yeast are an ideal system to study Heat Shock Protein 70 (Hsp70) function in a cellular context. This protocol was generated to analyze the function of non-native Hsp70 proteins by expressing them as the sole cytosolic Hsp70 in yeast. As an initial step, Hsp70 variants (such as Ssa1 point mutants and non-yeast versions such as Nematostella vectensis NvHsp70A, B and D) are cloned into an appropriate expression plasmid. Next, these plasmids are transformed into ssa1-4∆ yeast [expressing native Ssa1 from an uracil-based (URA3) plasmid] which are subsequently cured of the original yeast on 5-Fluroorotic Acid (5-FOA). The resulting cells can be screened for a variety of phenotypes which match to the activity of well-studied cellular pathways.
Keywords: Hsp70Background
cHsp70 is a molecular chaperone that plays a role in protein folding of newly synthesized and misfolded proteins (Rosenzweig et al., 2019). It also controls the activity of regulatory proteins that contribute to cell cycle progression, protein degradation, apoptosis and resistance to anticancer therapeutics. Hsp70 is highly conserved throughout nature and is essential for cell viability. Organisms can express several highly similar Hsp70 isoforms. For example, the budding yeast Saccharomyces cerevisiae expresses 4 cytosolic isoforms, Ssa1-4 (Lotz et al., 2019). In order to characterize Hsp70 function, we can express different Hsp70 isoforms, paralogs, point mutations and truncations as the sole cytosolic Hsp70 protein in S. cerevisiae using a 5-FOA plasmid swap strategy (Boeke et al.,1987).
We were recently able to use this methodology to express Nematostella vectensis Hsp70 isoforms (NvHsp70A, B and D) in yeast and asses their functionality through phenotypic screens, growth assays and analysis of interaction partners (Waller et al., 2018; Knighton et al., 2019). These results contribute to a better understanding of Hsp70 isoform function and the potential basis of local adaptation in populations of N. vectensis.
Materials and Reagents
- Pipette tips (10 μl, 300 μl, 1000 μl) (Neptune, catalog number: 2347; US Scientifica, catalog number: 1110-9700; Neptune, catalog number: 2167)
- Eppendorf 1.5 ml tubes (VWR, catalog number: 87003-294)
- 15 ml Falcon tubes (VWR, catalog number: 525-0449)
- 50 ml Falcon tubes (VWR, catalog number: 525-0447)
- Petri dishes (Thermo Fisher, catalog number: FB0875713)
- 96-well microplate (Greiner bio-one, catalog number: 655101)
- pAG415GPD-ccdB vector (Addgene plasmid # 14146; http://n2t.net/addgene:14146; RRID: Addgene_14146)
- Q5® High-Fidelity 2x Master Mix (NEB, catalog number: M0492S)
- ssa1-4∆ yeast strain (Jaiswal et al., 2011)
- Top 10 E. coli cells (Invitrogen, catalog number: C404003)
- Single-stranded carrier DNA (salmon sperm DNA, Solarbio, catalog number: D8030)
- Tryptone (US Biological Life Sciences, catalog number: C16050360)
- Ampicillin (Thermo Fisher, catalog number: 1159027)
- Yeast extract (US Biological Life Sciences, catalog number: C16091364)
- Peptone (HIMEDIA, catalog number: RM001)
- Glucose (VWR Amresco Life Science, catalog number: 0188)
- Agar (IBS Scientific, catalog number: IB49171)
- Adenine hemisulfate salt (Acros Organics, catalog number: 163631000)
- Yeast nitrogenous base (US Biological Life Sciences, catalog number: C16121501)
- Drop out mix minus leucine, methionine, uracil (US Biological Life Sciences, catalog number: C15050101)
- Drop out mix minus leucine (US Biological Life Sciences, catalog number: C15121094)
- Lithium Acetate (LiAc) (Sinopharm Chemical Reagent, catalog number: 30109760)
- Polyethylene glycol (PEG) (Sigma-Aldrich, catalog number: P3640)
- 0.05 g uracil (Arcos organics, catalog number: XLE-1000)
- 5-FOA (US biological life sciences, catalog number: 16052511)
- Hydroxyurea (HU) (Chem-implex Int’L INC, catalog number: 24533)
- Methyl methanesulfonate MMS (Acos organics, catalog number: 156891000)
- Sodium chloride (NaCl) (VWR, amrescolife science, catalog number: 0188)
- Cadmium chloride (CdCl2) (Acros organics, catalog number: 42350500)
- Hydrogen peroxide (H2O2) (Sigma-Aldrich, catalog number: 216763)
- Copper Chloride (CuCl2) (Sigma-Aldrich, catalog number: 222011)
- 2 ml of methionine (1 g/100 ml) (Arcos organics, catalog number: AC166160250) (see Recipes)
- YPDA Media (1 L) (see Recipes)
- Synthetic drop-out plates/media (1 L) (see Recipes)
- 5-FOA media (1 L) (see Recipes)
- Stock solution of 100 mM CuCl2 (see Recipes)
- Stock solution of CdCl2 (see Recipes)
- Chemical plates (see Recipes)
- 1 M LiAc (500 ml) (see Recipes)
- 100 mM LiAc (500 ml) (see Recipes)
- 50% (w/v) PEG (500 ml) (see Recipes)
Equipment
- UV radiator (Spectroline UV Crosslinker, model: XLE-1000)
- 30 °C Incubator (VWR, catalog number: 89511-422)
- 37 °C incubator (VWR, catalog number: 89511-428)
- Heraeus Pico 21 Microcentrifuge (Thermo Fisher scientific, catalog number: 75002416)
- Heraeus Multifuge X1 Centrifuge Series (Thermo Fisher scientific, catalog number: 75004211)
- 30 °C Incubator Shaker (New Brunswick Excella E25)
- Thermomixer (Eppendorf, catalog number: 5382EJ909137)
- Heat block (Thermo Scientific, catalog number: 88870000)
- Thermal cycler (Bio-Rad, catalog number: 621BR26941)
- Replica plater for 96-well plate 8X6 Array (Sigma-Aldrich, catalog number: R2383-1EA)
- Autoclave (Steris Amsco Cnetury, catalog number: SG-120)
- pH reader (RPI, catalog number: 850063)
Procedure
- Cloning of Nematostella vectensis Hsp70 isoforms into yeast expression plasmids
- There are three Nematostella vectensis Hsp70 cytosolic isoforms; NvHsp70A, NvHsp70B, NvHsp70D.
- We assembled the open reading frame from each Hsp70 (A, B, D) using sequence resources available through Nematostella JGI genome portal (https://genome.jgi.doe.gov/portal/).
- NvHsp70A, B, and D were amplified from cDNA synthesized from RNA isolated from Nematostella vectensis originating from Massachusetts coast.
- The published protocol for Q5® High-Fidelity 2x Master Mix (https://www.neb.com/protocols) was used with an annealing temperature of 60 °C and extension time of 1 min.
- The following primers were used (Table 1):
Table 1. Primer sequences used for cloning NvHsp70 isoforms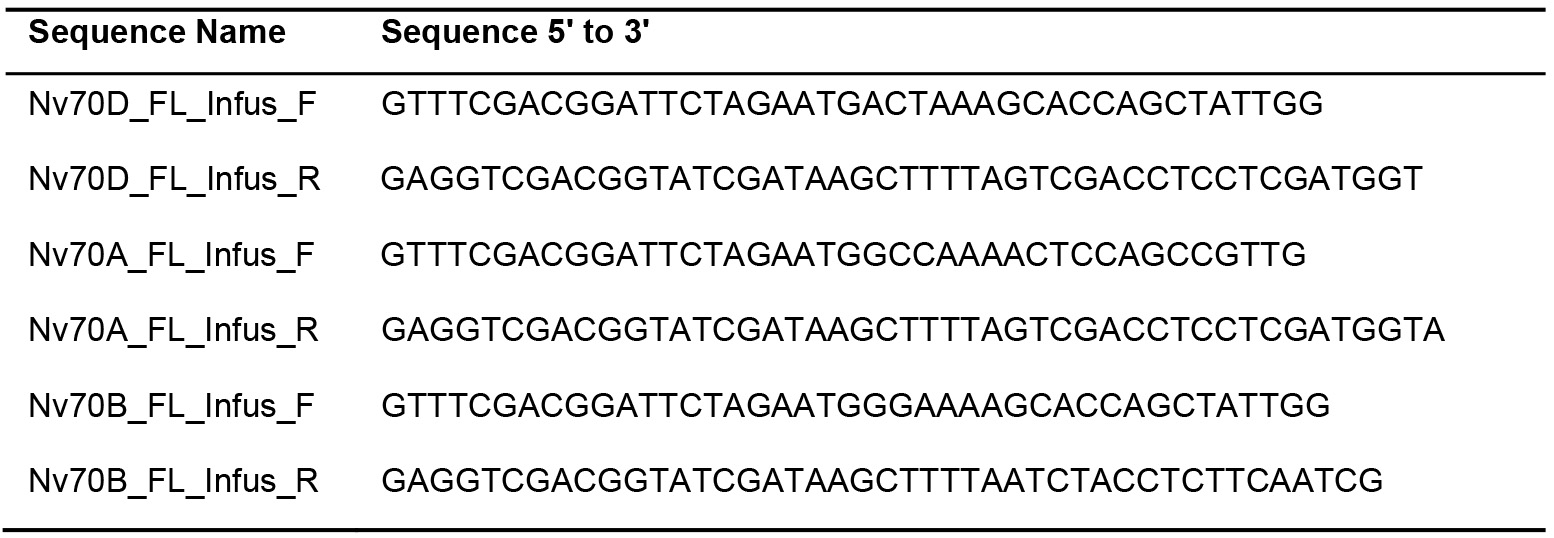
- Infusion cloning was used to integrate PCR products into yeast expression plasmid pAG415GPD-ccdB vector
- The main yeast cytosolic Hsp70, Ssa1 was used as a control.
- All inserts were sequence confirmed with Sanger sequencing.
- Ssa1-4∆ yeast strain
S. cerevisiae contains four isoforms of cytosolic Hsp70 (Ssa1-4), at least one of the isoforms is required in order to remain viable. The ssa1∆ ssa2∆ ssa3∆ ssa4∆ (ssa1-4∆) pYCPlac33-SSA1 strain was generated using the loxP-kanMX-loxP gene disruption cassette. In order to keep the ssa1-4∆ strain, a plasmid-borne pYCPlac33-SSA1 (URA3) is needed. For more strain details, please see (Jaiswal et al., 2011). - Transformation of Hsp70 expression plasmids into ssa1-4∆ yeast cells
- Inoculate 1 colony in 10 ml of YPD (See Recipes) and incubate in a shaker overnight at 30 °C.
- The following morning, inoculate 100 ml of YPD with the 10 ml overnight culture and incubate at 30 °C (shaking at 220 rpm) until an OD600nm of 0.5 is achieved.
- Harvest the culture in a sterile 50 ml centrifuge tube at 3,000 x g for 5 min.
- Pour off medium, re-suspend the cells in 25 ml of sterile H2O and centrifuge again.
- Pour off the H2O, re-suspend the cells in 1.0 ml of 100 mM LiAc and transfer the suspension to a sterile 1.5 ml microfuge tube.
- Pellet the cells at top speed for 5 s and remove the LiAc with a micropipette.
- Re-suspend the cells to a final volume of 500 μl which is about 400 μl of 100 mM LiAc.
- Boil a 1.0 ml sample of single stranded carrier DNA for 5 min and quickly chill in ice water.
- Vortex the cell suspension and pipette 50 μl samples into labeled microfuge tubes. Pellet the cells and remove the LiAc with a micropipette.
- The basic “transformation mix” consists of the following ingredients; carefully add them in order listed:
- 240 μl of PEG (50%w/v)
- 36 μl of 1.0 M LiAc
- 25 μl of single stranded carrier DNA (2.0 mg/ml)
- 50 μl of H2O and cloned NvHsp70 plasmid DNA (0.1-10 μg)
- Vortex each tube vigorously until the cell pellet has been completely mixed. This usually takes about one minute.
- Incubate the cell suspension for 30 min at 30 °C shaking at 220 rpm.
- Heat shock the cell suspension for 20-25 min in a water bath at 42 °C.
- Incubate on ice for 2 min.
- Microfuge at 5,000 x g for 15 s and remove the transformation mix with a micropipette.
- Pipette 0.2-1.0 ml of sterile H2O into each tube and re-suspend the pellet by pipetting it up and down gently.
- Screening the positive yeast colonies on selective media
- Warm and dry appropriate plates in a 30 °C incubator for at least 30 min prior to plating cells.
- After transformation, spread the yeast cells on synthetic dropout media (SD, Recipe 2) supplemented with the appropriate nutrients to select for plasmids and incubate at 30 °C for 2 days.
- Streak the colonies to fresh SD plates and incubate at 30 °C for at least 1 day.
- Streak yeast cells from previous to SD media containing 5-fluoro-orotic acid (5-FOA, Recipe 3) to counter-select for the URA3-based covering vector.
- If the mutated or non-native Hsp70 is insufficient to keep the cells alive as the sole isoform, the cells will not grow on plates containing 5-FOA.
- At this point, cells may be re-streaked and kept on YPD media. It is not required to keep them on SD-Leu as the Hsp70 plasmid is essential for viability.
- Testing functionality of Hsp70 under different treatments
- If the non-native Hsp70 is able to provide essential function as the sole Hsp70 isoform of the cell at normal conditions (30 °C), we can further characterize the functionality of Hsp70 by observing the cellular resistance to different stressors including heat stress (37 °C), DNA damage (Hydroxyurea), Oxidative stress (H2O2), NaCl (osmotic stress), UV radiation, and CdCl2 and CuCl2 (heavy metal exposure).
- Ssa1-4∆ cells expressing NvHsp70 isoforms as their sole cytosolic Hsp70 are grown to mid-logarithmic phase (OD600nm = 0.5) in 10 ml of YPDA in a 50 ml tube.
- Dilute cells 10-fold serially (full concentration, 1/10 concentration, 1/100 concentration, and 1/1,000 concentration) in a clear 96-well plate.
- Cells are replica plated onto solid YPDA media containing the aforementioned chemicals or normal YPDA media and then exposed to abiotic stressors including heat stress at 37 °C and UV radiation (150 Jm-2, 200 Jm-2).
- Concentration of drugs used are as follows; HU (200 mM and 300 mM), MMS (0.04%, 0.08%), NaCl (0.8 M,1 M), CdCl2 (30 mM, 45 mM), CuCl2 (3 mM, 4 mM, 5 mM), H2O2 (0.8 mM, 1 mM).
- After cells have dried, all plates (except for the 37 °C plate) are kept upside-down in a 30 °C incubator for 3 days.
- A control plate of cells replica plates on normal YPDA (kept at 30 °C) should be used to compare the “normal growth rate” of the non-native isoforms in yeast to that on the chemical plates, see Figure 1.

Figure 1. Expressing NvHsp70 in yeast (Waller et al., 2018). NvHsp70 isoforms provide essential Hsp70 function in yeast. Ssa1–4∆ cells were transformed with control plasmid pAG415-ccDB, Ssa1-expressing, or NvHSP70 isoform-expressing plasmids and then serially diluted onto media lacking leucine or containing 5-FOA. Growth of cells on 5-FOA demonstrates the ability of NvHSP70 isoforms to provide essential function when expressed as the sole Hsp70 in the cell. Plates were incubated for 3 days at 30 °C and then photographed.
Data analysis
Photographs of the plates should be taken after a pattern of cell growth can be observed, typically after 3 days (see Figure 1).
Notes
Although this protocol describes the expression and analysis of Hsp70 isoforms from Nematostella vectensis, it can also be used to analyze Hsp70 phosphorylation site function as in (Truman et al., 2012) or allow analysis of global interactions of Hsp70 using epitope tagged versions as in (Truman et al., 2015a and 2015b; Knighton et al., 2019)
Recipes
- 50% (w/v) PEG (500 ml)
Dissolve 250g of polyethylene glycol in 300 ml of ddH2O
Add ddH2O until final volume is 500 ml
Autoclave at 121.0 °C on 1-hour liquid cycle - 1 M LiAc (500 ml)
Dissolve 51 g of Lithium Acetate in 300 ml of ddH2O
Add ddH2O until final volume is 500 ml
pH to 7.5
Autoclave at 121.0 °C on 1-hour liquid cycle - 100 mM LiAc (500 ml)
Dilute 1 M LiAc by 10x
Autoclave at 121.0 °C on 1-hour liquid cycle - YPDA Media (1 L)
800 ml ddH2O
10 g Yeast Extract
20 g Peptone
20 g glucose
40 mg of Adenine
Add 20 g of Agar (for plates only)
Add ddH2O to make the volume up to 1 L
Pour into appropriate size bottles
Autoclave at 121.0 °C on 1-hour liquid cycle
Let cool and dry overnight at room temperature - Synthetic drop-out plates/media (1 L)
800 ml ddH2O
6.7 g YNB
20 g glucose
1.62 g Dropout mix -leu
0.05 g of adenine
Adjust the pH to 6.0 with sodium hydroxide (if lower than 6)
Add 20 g of Agar (for plates only)
Add RO water to get to 1 L
Pour into appropriate size bottles
Autoclave at 121.0 °C on 1-hour liquid cycle
Let cool and dry overnight at room temperature - 5-FOA media (1 L)
6.7 g YNB
20 g glucose
1.47 g of -ura, -leu, -met dropout
0.05 g uracil
2 ml of methionine
Add 20 g agar and make up to 1 L
Autoclave at 121.0 °C on 1-hour liquid cycle
After autoclaving, let it cool slightly while stirring and then add 1 g of FOA
Pour plates
Let cool and dry overnight at room temperature - Stock solution of 100 mM CuCl2
Measure 0.166 g of CuCl2 and mix with 15 ml of water - Stock solution of CdCl2
Measure 1.67 g of CdCl2 and mix with 15 ml of water - Chemical plates
Hydroxyurea (HU)- Measure 0.76 g of HU and mix gently with 50 ml of liquid YPDA (cool to the touch) and pour into two Petri dishes.
- Let cool and dry overnight at room temperature
- Measure 0.02 ml of MMS and mix gently with 50 ml of liquid YPDA (cool to the touch) and pour into two Petri dishes to make 0.04% MMS
- Measure 0.04 ml of MMS and mix gently with 50 ml of liquid YPDA (cool to the touch) and pour into two Petri dishes to make 0.08% MMS
- Let cool and dry overnight at room temperature
CuCl2- From 100 mM CuCl2 stock solution measure out 1.5 μl and mix with 50 ml of liquid YPDA (cool to the touch) to make a 3.0 mM concentration and pour into two Petri dishes
- From 100 mM CuCl2 stock solution measure out 2 μl and mix with 50 ml of liquid YPDA (cool to the touch) to make a 4.0 mM concentration and pour into two Petri dishes
- From 100 mM CuCl2 stock solution measure out 2.5 μl and mix with 50 ml of liquid YPDA (cool to the touch) to make a 5.0 mM concentration and pour into two Petri dishes
- Let cool and dry overnight at room temperature
CdCl2- From the CdCl2 stock solution measure out 15 μl and mix with 50 ml of liquid YPDA (cool to the touch) and pour into Petri dishes
- From the CdCl2 stock solution measure out 22.5 μl and mix with 50 ml of liquid YPDA (cool to the touch) and pour into Petri dishes
- Let cool and dry overnight at room temperature
- Add 40 μl of H2O2 and mix with 50 ml of YPDA (cool to the touch) to make 0.8 mM concentration and pour into two Petri dishes. And let it cool overnight
- Add 50 μl of H2O2 and mix with 50 ml of YPDA (cool to the touch) to make 1 mM concentration and pour into two Petri dishes. And let it cool overnight
- Add 75 μl of H2O2 and mix with 50 ml of YPDA (cool to the touch) to make 1.5 mM concentration and pour into two Petri dishes. And let it cool overnight
- Let cool and dry overnight at room temperature
- Measure 2.34 g NaCl and mix with 50 ml of YPDA (cool to the touch) to make a 0.8 M concentration and pour into two Petri dishes
- Measure 2.92 g NaCl and mix with 50 m of YPDA (cool to the touch) to make a 1.0 M concentration and pour into two Petri dishes
- Let cool and dry overnight at room temperature
Acknowledgments
This research was supported by the National Institutes of Health NCI R15CA208773 (AWT) and National Science Foundation NSF DEB1545539 and REU #1359271 (AMR). We thank our colleagues in both the Truman and Reitzel labs who provided assistance, insight, and comments that greatly assisted both the research and the manuscript. This protocol was adapted from our previous work (Waller et al., 2018).
Competing interests
The authors of this manuscript declare no competing interests.
References
- Boeke, J. D., Trueheart, J, Natsoulis, G. and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Method Enzymol 154:164-175.
- Jaiswal, H., Conz, C., Otto, H., Wolfle, T., Fitzke, E., Mayer, M. P. and Rospert, S. (2011). The chaperone network connected to human ribosome-associated complex. Mol Cell Biol 31(6): 1160-1173.
- Knighton, L. E., Nitika, Waller, S. J., Strom, O., Wolfgeher, D., Reitzel, A. M. and Truman, A. W. (2019). Dynamic remodeling of the interactomes of Nematostella vectensis Hsp70 isoforms under heat shock. J Proteomics 206: 103416.
- Lotz, S. K., Knighton, L. E., Nitika, Jones, G. W. and Truman, A. W. (2019). Not quite the SSAme: unique roles for the yeast cytosolic Hsp70s. Curr Genet 65(5):1127-1134.
- Rosenzweig, R., Nillegoda, N. B., Mayer, M. P. and Bukau, B. (2019). The Hsp70 chaperone network. Nat Rev Mol Cell Biol. doi: 10.1038/s41580-019-0133-3.
- Truman, A. W., Kristjansdottir, K., Wolfgeher, D., Hasin, N., Polier, S., Zhang, H., Perrett, S., Prodromou, C., Jones, G. W. and Kron, S. J. (2012). CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 151(6): 1308-1318.
- Truman, A. W., Kristjansdottir, K., Wolfgeher, D., Ricco, N., Mayampurath, A., Volchenboum, S. L., Clotet, J. and Kron, S. J. (2015a). Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase. J Proteomics 112: 285-300.
- Truman, A. W., Kristjansdottir, K., Wolfgeher, D., Ricco, N., Mayampurath, A., Volchenboum, S. L., Clotet, J. and Kron, S. J. (2015b). The quantitative changes in the yeast Hsp70 and Hsp90 interactomes upon DNA damage. Data Brief 2: 12-15.
- Waller, S. J., Knighton, L. E., Crabtree, L. M., Perkins, A. L., Reitzel, A. M. and Truman, A. W. (2018). Characterizing functional differences in sea anemone Hsp70 isoforms using budding yeast. Cell Stress Chaperones 23(5): 933-941.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Knighton, L. E., Saa, L. P., Reitzel, A. M. and Truman, A. W. (2019). Analyzing the Functionality of Non-native Hsp70 Proteins in Saccharomyces cerevisiae. Bio-protocol 9(19): e3389. DOI: 10.21769/BioProtoc.3389.
Category
Microbiology > Heterologous expression system > Saccharomyces cerevisiae
Molecular Biology > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








