- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Highly Sensitive, Reproducible Assay for Determining 4-hydroxynonenal Protein Adducts in Biological Material
Published: Vol 9, Iss 19, Oct 5, 2019 DOI: 10.21769/BioProtoc.3383 Views: 3957
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Determining Ribosome Translational Status by Ribo-ELISA
Amandine Bastide [...] Alexandre David
Jan 5, 2018 7874 Views

Protocol for RYMV Inoculation and Resistance Evaluation in Rice Seedlings
Agnès Pinel-Galzi [...] Laurence Albar
Jun 5, 2018 8062 Views

An Optimised Indirect ELISA Protocol for Detection and Quantification of Anti-viral Antibodies in Human Plasma or Serum: A Case Study Using SARS-CoV-2
Claire Baine [...] Jennifer Serwanga
Dec 20, 2023 3637 Views
Abstract
Oxidative stress is associated with numerous diseases, and markers of oxidative stress in biological material are becoming a mainstay of both experimental and clinical/epidemiological research. Lipid peroxidation is a major form of oxidative stress, but due to their rapid degradation and instability, lipid peroxides are notoriously difficult to measure, particularly in biological specimens where their production and removal are continuously occuring. Thus, a commonly used surrogate marker of lipid peroxidation is protein adducts of 4-Hydroxynonenal (HNE), an α, β-unsaturated hydroxyalkenal (i.e., a reactive aldehyde) formed via degradation of oxidized polyunsaturated fatty acids (PUFAs). HNE adducts can be measured via commercially-available immunosorbent assays, but these have their limitations due to excessive costs, and reproducibility among laboratories is challenging due to variability in assay sensitivity, procedure, and reagents. Here we present a reproducible, facile, and economically conservative protocol for quantifying HNE protein adducts. The key to this protocol is to generate HNE-adduct standards by incubating bovine serum albumin (BSA) with HNE. These standards are then adsorbed to immunsorbent plastic in a multi-well plate format alongside biological samples. An enzyme-linked immunosorbent assay (ELISA) is then performed on the multi-well plate using commercially-available primary and secondary antibodies, and a peroxide-based fluorescent developing reagent. This protocol is highly sensitive and offers advantages to commercial sources in that it allows for reproducible, high-throughput quantitation of HNE adducts in a large number of samples.
Keywords: Oxidative stressBackground
When polyunsaturated fatty acids (PUFAs) become oxidized in cell membranes, there are only two possible outcomes: 1) resulting lipid peroxides (LOOHs) are neutralized via enzyme-dependent (e.g., glutathione peroxidase-4, GPx4) and/or spontaneous reaction with redox-active molecules (e.g., quinones); or 2) the LOOHs degrade into highly reactive aldehydes, capable of forming adducts with proteins, DNA and other lipids. One of the more common aldehydes formed is 4-hydroxynonenal (HNE), an α, β-unsaturated aldehyde formed from peroxidation of n-6 PUFAs such as arachidonic acid and linoleic acid, both of which are abundant in phospholipid cell membranes. Accumulation of HNE and other related biogenic aldehydes, such as malondialdehyde, react with nucleophilic side chains in proteins and polypeptides to form stable protein adducts that can sometimes act as ‘toxic second messengers’ of oxidative stress. These reactions constitute a form of oxidative stress referred to as carbonyl stress. Hydroxynonenal can noncompetitively inhibit the activity of antioxidant enzymes like aldehyde dehydrogenase (ALDH2) and aldo-ketoreductase (AKR), further exacerbating accumulation and cytotoxicity of reactive carbonyl species (Jinsmaa, et al., 2009). Since LOOHs are notoriously difficult to directly measure in vivo, HNE-adducts serve as a surrogate biomarker of LOOHs, and the corresponding carbonyl stress imposed by them in cells and tissues have been reported in both experimental models and clinical studies of degenerative and age-related pathologies including Parkinson’s Disease, obesity and diabetes, cardiovascular diseases, and many cancers (Markesbery and Lovell, 1998; Orioli et al.; 1998; Selley, 1998; Traverso et al., 1998; Frohnert et al., 2011; Anderson et al., 2014; Katunga et al., 2015a; Anderson et al., 2018). Hydroxynonenal tends to react with nucleophilic moieties, such as the side-chains of cysteine, lysine, and histidine, either the carbonyl group, forming a Schiff base, or the β-carbon, forming a Michael addition adduct.
A variety of assays for HNE-adduct detection are commercially available. Most of them employ an ELISA method, using a proprietary combination of antigen-capture or ‘sandwich-ELISA’ formulation. These assays can vary widely in sensitivity, leading to problems with reproducibility and data compatibility across laboratories. Here we present a facile, economically conservative and reproducible assay for determining HNE-adduct levels in serum, cell-, and tissue-lysates.
Materials and Reagents
- Paper towel
- 96-Well Plate (Clear Flat-Bottom, MaxiSorp, Thermo Fischer Scientific, catalog number: 442404)
- 4-hydroxynonenal (Caymen Chemical, catalog number: 32100)
- Bovine Serum Albumin solution, sterile (Sigma-Aldrich, catalog number: A9576)
- Immuno Shot-Platinum (Cosmo Bio, catalog number: CSR-IS-P-500)
- Anti-HNE antibody (Percipio Biosciences, catalog number: 24327)
- Goat Anti-Mouse HRP conjugated antibody (Santa Cruz Bio Technology, catalog number: 170-5047)
- Amplex UtraRed Reagent (Thermo Fischer Scientific, catalog number: A36006)
- Sodium Phosphate Buffer (50 mM, pH 7.4)
- NaCl (Sigma-Aldrich, catalog number: S9888)
- KCl (Sigma-Aldrich, catalog number: P3911)
- Na2HPO4·2H2O (Sigma-Aldrich, catalog number: 71643)
- KH2PO4 (Sigma-Aldrich, catalog number: 795488)
- Tween 20 (Sigma-Aldrich, catalog number: P1379)
- RIPA Buffer (Thermo Fischer, catalog number: 89901)
- Tris Base (Roche, catalog number: TRIS-RO)
- EDTA (Sigma-Aldrich, catalog number: EDS)
- EGTA (Sigma-Aldrich, catalog number: E3889)
- Triton X-100 (Sigma-Aldrich, catalog number: X100)
- H2O2
- 1x Phosphate Buffered Saline (PBS, see Recipes)
- 1x ELISA Wash Buffer (see Recipes)
- 1x Amplex UltraRed Solution (see Recipes)
- 1x TEE-T Buffer (see Recipes)
Equipment
- Incu-Mixer (Benchtop Scientific, model: MP4)
- Standard Lab Rocker (any brand)
- Fluorescent multi-well plate reader
- Duall Tissue Grinder (Kimble, model: 3432N07)
Procedure
- Sample Preparation
- Serum
Dilute serum 1:10 in PBS (see Recipes) before plating. - Tissue Homogenate
Using a Duall Tissue Grinder (Kimble) and a 1 g: 10 ml weight to volume ratio, homogenize heart tissue in TEE-T buffer (see Recipes). - Cell Lysate
Lyse cells using RIPA buffer at a ratio of 1 ml buffer: 106 cells. - Positive Controls
For cell culture: Add 100 μM H2O2 to cells for 2 h, and prepare cell lysate in RIPA buffer. Include those samples on ELISA plate.
For serum or tissue homogenate: After homogenization and prior to antigen adsorption step (Step 2), add 1.25 μM HNE-BSA adduct (from Step 2) to at least 2 unknown samples, and include those samples on ELISA plate.
- Serum
- Preparations of HNE-adduct solutions for standard curve
- A standard curve is prepared fresh, one day prior to each ELISA, with serial dilutions of HNE in 1% BSA (diluting BSA solution in 50 mM PBS, pH 7.4). While keeping BSA constant, HNE is varied serially (0-0.019-0.0375-0.075-0.15-0.31-0.625-1.25-2.5-5 μM) in a 200 μl solution of 1% BSA.
- Incubate at 37 °C for 24 h.
- Antigen adsorption
- Add 100 μl of standard curve solutions and 100 μl of unknown samples to wells, in triplicate, and cover with adhesive seal.
- Incubate overnight at 4 °C while rocking.
- Plate wash
- Empty plate and remove excess fluid by blotting on paper towel.
- Add 200 μl of ELISA wash buffer to each well.
- Repeat Steps 4a and 4b until the plate has been washed a total of 6 times.
- Blocking
- Add 200 μl of Immuno Shot-Platinum blocking agent to each well and cover with adhesive seal.
- Incubate for 4 h at room temperature while rocking.
- Plate wash
Wash plate as described in Step 4. - Primary antibody incubation
- Add 100 μl of a 1:100 dilution of primary antibody (Percipio Biosciences) in PBS to each well and cover with adhesive seal.
- Incubate for 2 h at 37 °C with agitation in the Incu-Mixer.
- Plate wash
Wash plate as described in Step 4. - Secondary antibody incubation
- Add 100 μl of a 1:7,500 dilution of secondary antibody (Santa Cruz BioTechnology,) in PBS to each well and cover with adhesive seal.
- Incubate for 2 h at 37 °C with agitation in the Incu-Mixer.
- Plate wash
Wash plate as described in Step 4. - Developing and reading fluorescence (Amplex UltraRed Solution)
- Add 100 μl of Amplex UltraRed Solution (prepared fresh) to each well and cover with adhesive seal.
- Incubate at room temperature, protecting from light.
- Using a multi-well plate reader, measure fluorescence in each well at ~560 nm/590 nm (ex/em spectrum for Resorufina, the oxidized product formed in the reaction of H2O2 with Amplex Red, catalyzed by HRP). Do this 15, 30, and 60 min after addition of Amplex UltraRed Solution.
Data analysis
- Enter fluorescent values for each sample into analytical software (e.g., Excel, Graphpad Prism, SigmaPlot).
- Fluorescent values for each time-point (15, 30, 60 min) must be compared with the values for the HNE-BSA standards at the same time-point. In some cases, 15 min may not be long enough, or 60 min may be too long. This needs to be individually optimized for each laboratory, and may vary based on sensitivity and availability of fluorescent plate-reader, and/or other factors.
- Standard curve is fit to proper equation (typically 3rd order polynomial).
- Values in the unknown sample wells are interpolated using equation for standard curve.
- Concentration of HNE-adducts in the unknown sample wells should be normalized to the protein concentration in each well.
Results
A representative HNE-adduct ELISA from our laboratory using tissue lysate prepared from human atrial biopsy is shown in Figure 1, and has previously been published (Katunga et al., 2015b). Lysate is prepared using TEE-T buffer as described in the Recipes below. The interpolated data shown in Figure 1C is then normalized to the protein concentration for each sample. As is clear from these data, sensitivity of the assay scales to the low nanomolar range for tissue lysate, but these values must be normalized to protein concentration within the well for final presentation. Final units are typically expressed as “nmol HNE-adduct/mg protein (or μmol/g).”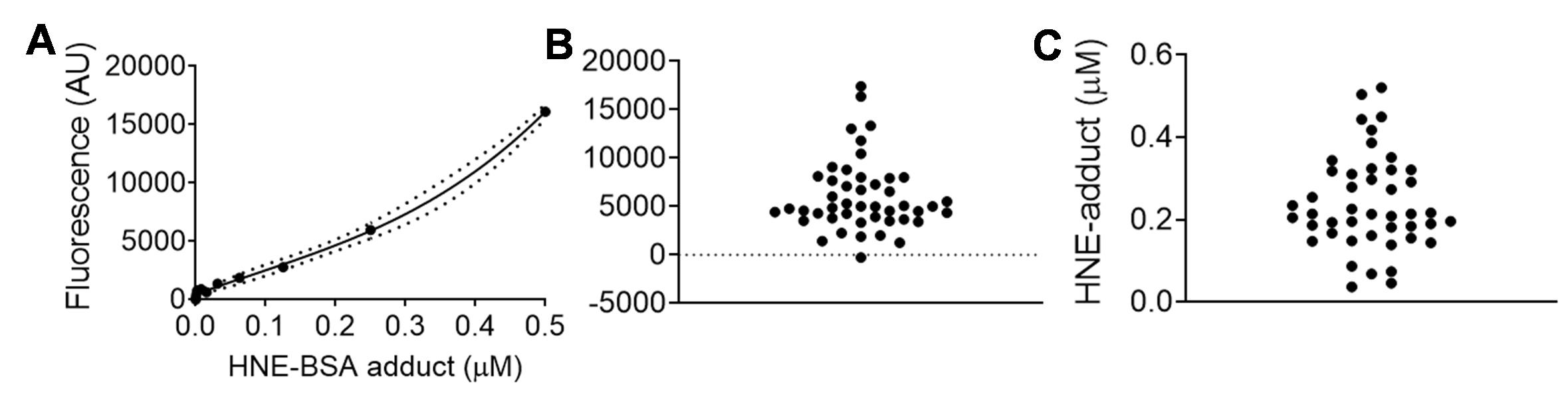
Figure 1. Representative data set of HNE-adduct ELISA used to measure HNE-adducts in human cardiac tissue homogenate/lysate prepared from right atrial appendage biopsy obtained at time of surgery (as described in Anderson et al., 2014; Katunga et al., 2015a). Samples were prepared as described in the protocol, and diluted 1:10 into PBS. In panel A is the standard curve of known concentrations of HNE-BSA adducts, in panel B is raw fluorescent data captured from samples on the same plate, and in panel C is the interpolated data from same samples. The standard curve was fitted using a third order polynomial equation (cubic), with 95% confidence intervals shown in dashed lines.
Recipes
- 1x Phosphate Buffered Saline (1 L, pH 7.4)
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4·2H2O
0.24 g KH2PO4 - 1x ELISA Wash Buffer (1 L)
0.5 ml of Tween 20
999.5 ml of PBS - 1x Amplex UltraRed Solution (10 ml, for full 96-well plate)
50 μl of Amplex UltraRed (10 mM in DMSO)
23 μl of 3% H2O2
9.925 ml of PBS
Note: Prepare immediately prior to Step 11. - 1x TEE-T Buffer (1 L, pH 7.5)
1.21 g Tris base
292 mg EDTA
380 mg EGTA
5 ml Triton-X
995 ml distilled water
Acknowledgments
This work was supported by funding from National Institutes of Health (HL122863, AG057006) and Department of Defense (PR181276) to E.J.A.
Competing interests
Authors have no conflicts of interest to disclose.
References
- Anderson, E. J., Thayne, K. A., Harris, M., Shaikh, S. R., Darden, T. M., Lark, D.S., Williams, J. M., Chitwood, W. R., Kypson, A. P. and Rodriguez, E. (2014). Do fish oil omega-3 fatty acids enhance antioxidant capacity and mitochondrial fatty acid oxidation in human atrial myocardium via PPARgamma activation? Antioxid Redox Signal. 21(8):1156-1163.
- Anderson, E. J., Vistoli, G., Katunga, L. A., Funai, K., Regazzoni, L., Monroe, T. B., Gilardoni, E., Cannizzaro, L., Colzani, M., De Maddis, D., Rossoni, G., Canevotti, R., Gagliardi, S., Carini, M. and Aldini, G. (2018). A carnosine analog mitigates metabolic disorders of obesity by reducing carbonyl stress. J Clin Invest 128(12): 5279-5293.
- Frohnert, B. I., Sinaiko, A. R., Serrot, F. J., Foncea, R. E., Moran, A., Ikramuddin, S., Choudry, U. and Bernlohr, D. A. (2011). Increased adipose protein carbonylation in human obesity. Obesity (Silver Spring) 19(9):1735-1741.
- Jinsmaa, Y, Florang, V. R., Rees, J. N., Anderson D.G., Strack, S. and Doorn, J.A. (2009) Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem Res Toxicol 22(5):835-841.
- Katunga, L. A., Gudimella, P., Efird, J. T., Abernathy, S., Mattox, T. A., Beatty, C., Darden, T. M., Thayne, K. A., Alwair, H., Kypson, A. P., Virag, J. A. and Anderson, E. J. (2015a). Obesity in a model of gpx4 haploins ufficiency uncovers a causal role for lipid-derived aldehydes in human metabolic disease and cardiomyopathy. Mol Metab 4(6):493-506.
- Katunga, L. A., Gudimella, P., Efird, J. T., Abernathy, S., Mattox, T. A., Beatty C, Darden, T. M., Thayne, K. A., Alwair, H., Kypson, A. P., Virag, J. A. and Anderson, E. J. (2015b). Obesity in a model of gpx4 haploinsufficiency uncovers a causal role for lipid-derived aldehydes in human metabolic disease and cardiomyopathy. Mol Metab 4(10):753.
- Markesbery, W. R. and Lovell, M. A. (1998). Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer’s disease. Neurobiol Aging 19(1):33-36.
- Orioli, M., Aldini, G., Benfatto, M. C., Facino, R. M. and Carini M. (2007). HNE Michael Adducts to Histidine and Histidine-Containing Peptides as Biomarkers of Lipid-Derived Carbonyl Stress in Urines: LC− MS/MS Profiling in Zucker Obese Rats. Anal Chem 79(23):9174-9184.
- Selley, M. L. (1998). (E)-4-hydroxy-2-nonenal may be involved in the pathogenesis of Parkinson’s disease. Free Radic Biol Med 25 (2):169-174.
- Traverso, N., Menini, S., Cosso, L., Odetti, P., Albano, E., Pronzato, M. A. and Marinari, U. M. (1998). Immunological evidence for increased oxidative stress in diabetic rats. Diabetologia 41(3):265-270.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Monroe, T. B. and Anderson, E. J. (2019). A Highly Sensitive, Reproducible Assay for Determining 4-hydroxynonenal Protein Adducts in Biological Material. Bio-protocol 9(19): e3383. DOI: 10.21769/BioProtoc.3383.
Category
Biochemistry > Protein > Posttranslational modification
Biochemistry > Protein > Immunodetection > ELISA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








