- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Robust, One-step FRET Assay for Human Heparanase
Published: Vol 9, Iss 17, Sep 5, 2019 DOI: 10.21769/BioProtoc.3356 Views: 4660
Reviewed by: Vivien Jane Coulson-ThomasPaul Ross Wildermantarsis ferreira

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
![[2-<sup>3</sup>H]Mannose-labeling and Analysis of N-linked Oligosaccharides](https://en-cdn.bio-protocol.org/imageup/arcimg/20170718074630401.jpg?t=1770580069)
[2-3H]Mannose-labeling and Analysis of N-linked Oligosaccharides
Marina Shenkman [...] Gerardo Z. Lederkremer
Jul 20, 2017 7824 Views

Analysis of Lipid-linked Oligosaccharides Synthesized in vivo in Schizosaccharomyces pombe
Ayelen Valko [...] Cecilia D’Alessio
Sep 20, 2022 2228 Views

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Kristina Seiler [...] Mario P. Tschan
Jul 20, 2023 2519 Views
Abstract
Heparanase, an endo-β-D-glucuronidase, cleaves cell surface and extracellular matrix heparan sulfate (HS) chains at distinct sites and plays important biological roles including modulation of cell growth and metastasis. Although a number of different types of heparanase assays have been reported to date, most are labor intensive, complex and/or expensive to carry out. We reasoned that a simpler heparanase assay could be developed using heparin labeled with Dabcyl and EDANS as donor and acceptor fluorophores so as to generate a FRET signal. Our results show that a more robust heparanase assay could be developed based on the principle studied herein and more homogeneous preparation of heparin. Yet, the assay in its current form could be used for routine screening of potential inhibitors in a high-throughput manner as well as for studying heparanase activity expressed in tumors as well as biological fluids like plasma.
Keywords: HeparanaseBackground
Human heparanase is an endo-β-D-glucuronidase that cleaves heparan sulfate (HS) chains present in proteoglycan form on cell surfaces and in the extracellular matrix (Fairbanks et al., 1999; Kussie et al., 1999; Toyoshima and Nakajima, 1999; Dempsey et al., 2000; Sanderson et al., 2017). Although other sequences are also likely to be targeted by heparanase (Peterson and Liu, 2013), substrate specificity studies indicate that heparanase preferably cleaves the 1→4-inter-glycosidic bond between a glucuronic acid (GlcA) and glucosamine-N,6-disulfate (GlcNS6S) residues.
The importance of heparanase as a therapeutic target has led to the development of several biochemical and biophysical assays over the past 2 decades. Despite the availability of numerous assays, no particular assay appears to have been broadly used to understand heparanase biology, pharmacology, and drug discovery. The reported assays typically suffer from the involvement of multiple steps and incubation up to 24 h (Freeman and Parish, 1997; Behzad and Brenchley, 2003; Tsuchida et al., 2004; Enomoto et al., 2006; Hammond et al., 2010; Schiemann et al., 2012; Melo et al., 2015). An easier, one-step assay would greatly help deduce inhibitors, understand substrate specificity, elucidate the mechanism of action, and clarify enzymatic or non-enzymatic role in cellular systems. A more robust heparanase assay would be easier to implement (fewer steps, faster screening time, no immobilization, no post-assay signal development), which is adaptable to microplate format, enable assaying active heparanase in cellular media, and perhaps help monitor heparanase in vivo. We hypothesized fluorescence resonance energy transfer (FRET)-based assay may offer a one-step solution that can address several attributes. This documents, method of developing a FRET-based assay for active heparanase. Based on our work, FRET-enabled heparin chain could effectively serve as a substrate of heparanase, help detect an active enzyme in media, and to screen potential inhibitors.
Materials and Reagents
- 1 cm path length quartz cuvette (Hellma®, catalog number: Z801712)
- 96-well plates (Corning, catalog number: 3897)
- Wilmad-LabGlassTM NMR Tubes (Fisher Scientific, 16-800-554)
- 6-well tissue culture-treated plates (Cyto one, catalog number: CC7672-7506)
- Low-Retention Microcentrifuge Tubes (Fisher Scientific, catalog number: 02-681-331; Amber, catalog number: 05-402-31)
- FisherbrandTM Micro Stir Bars (Fisher Scientific, catalog number: 16800523)
- FLEX-COLUMN® Economy Columns (Fisher Scientific, catalog number: K420400-1030)
- SpectrumTM Spectra/PorTM Float-A-LyzerTM G2 Dialysis Devices (Fisher Scientific, catalog number: 08-607-020)
- Fisher brandTM Class B Amber Glass Threaded Vials with Attached Caps (Fisher Scientific, catalog number: 14-955-332)
- PierceTM Protein Concentrator PES, 3K MWCO, 2-6 ml (Fisher Scientific, catalog number: PI88515)
- Sephadex G-15 (GE Healthcare, catalog number: 17002001)
- MCF7 (ATCC® HTB-22TM) and HEK 293T (ATCC® ACS-4500TM) cell lines
- Heparin sodium salt from porcine intestinal mucosa (Sigma-Aldrich, catalog number: H4784-1G stored at 4 °C)
- Dabcyl C2 amine (100 mg) (AnaSpec, catalog number: AS-81819, stored at -20 °C under N2)
- EDANS, sodium salt, 5- [(2-aminoethyl) amino] naphthalene-1-sulfonic acid, sodium salt (0.5 g) (AnaSpec, catalog number: AS-23886, stored at -20 °C under N2)
- N-(3-Dimethylaminopropyl)-N′-ethyl carbodiimide hydrochloride (EDC) (Sigma-Aldrich, catalog number: 03449, stored at -20 °C)
- N-hydroxysuccinimide (NHS) (Sigma-Aldrich, catalog number: 130672-5G, -20 °C under N2 )
- Glacial Acetic Acid (HPLC grade) (Fisher Scientific, catalog number: A35-500)
- Methyl sulfoxide, 99.7%, Extra Dry, anhydrous, SC (AcroSealTM, ACROS OrganicsTM, catalog number: AC610971000)
- Thermo ScientificTM NERLTM High Purity Water (Fisher Scientific, catalog number: 23-249-590)
- Water (HPLC grade) (Fisher Scientific, Fisher Chemical W5-4)
- Suramin (Sigma-Aldrich, catalog number: S2671)
- Deuterium oxide, D2O (Sigma-Aldrich, catalog number: 151882)
- Sodium acetate (Sigma-Aldrich, catalog number: S7545)
- Sodium Chloride (Fisher Scientific, catalog number: BP358-1)
- HEPES (Fine White Crystals/Molecular Biology) (Fisher BioReagents, catalog number: BP310-500)
- HyCloneTM Fetal Bovine Serum (FBS) (Fisher BioReagents, catalog number: SH3007102)
- Dulbecco's Modified Eagle Medium (DMEM) growth media (Invitrogen, USA)
- (Optional) Heparanase protein (R&D systems, catalog number: 7570-GH-005)
- Antibiotic-antimycotic Solution (AA) (Fisher Scientific, catalog number: 15-240-062), antibiotic spectrum: Amphotericin B, Penicillin, Streptomycin
- Assay buffer (see Recipes)
Equipment
- FreeZone Plus 6 Lyophilizer (Labconco Corporation)
- Peristaltic Pump P-1 (GE healthcare, catalog number: 18111091 )
- Temperature controlled Centrifuge (Beckman coulter, model: Microfuge 20R; Eppendorf, model: 5804R)
- Flex Station III (Molecular Devices, CA)
- Shimadzu Spectrophotometer (Shimadzu Scientific Instruments, USA)
- Fluorimeter PTI QM-400 (Horiba Canada)
- pH meter (Denver Instrument, model: 220)
- 400 MHZ topspin H1NMR Bruker
- C24 classic series Incubator (New Brunswick Scientific, NJ)
- Precision Water bath (Precision Scientific, model: 25)
- -20 °C (Norlake Scientific) and -80 °C freezer (Thermo scientific)
- Forma Series II, water jacketed CO2 incubator (Thermo Electron Corporation, USA)
- Stirring Hotplate (FisherbrandTM IsotempTM, catalogue number: SP88850200)
- Fluorescence cuvette (Hellma®, catalog number: 101.015-QS; Cell Holder, catalog number: 013.013)
Software
- SigmaPlot 13 (Systat Software, Inc. or OriginPro or Kaleidagraph)
Procedure
- Heparin Labeling
- Weigh 100 mg Heparin and transfer into an amber glass vial.
- Dissolve in 0.1-0.2 ml distilled H2O.
- Add 5.9 mg N-hydroxysuccinimide (NHS) followed by 25.6 mg 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC). Stir mixture using a micro stir bar for 3 min.
- Weigh 4.1 and 3.8 mg Dabcyl C2 amine and EDANS respectively into an amber eppendorf and dissolve in 100 μl DMSO. Refer Table 1 for stoichiometrices.
Table 1. Stoichiometrices used for the synthesis of labeled Heparin (% Yield after size exclusion chromatography: 70-75)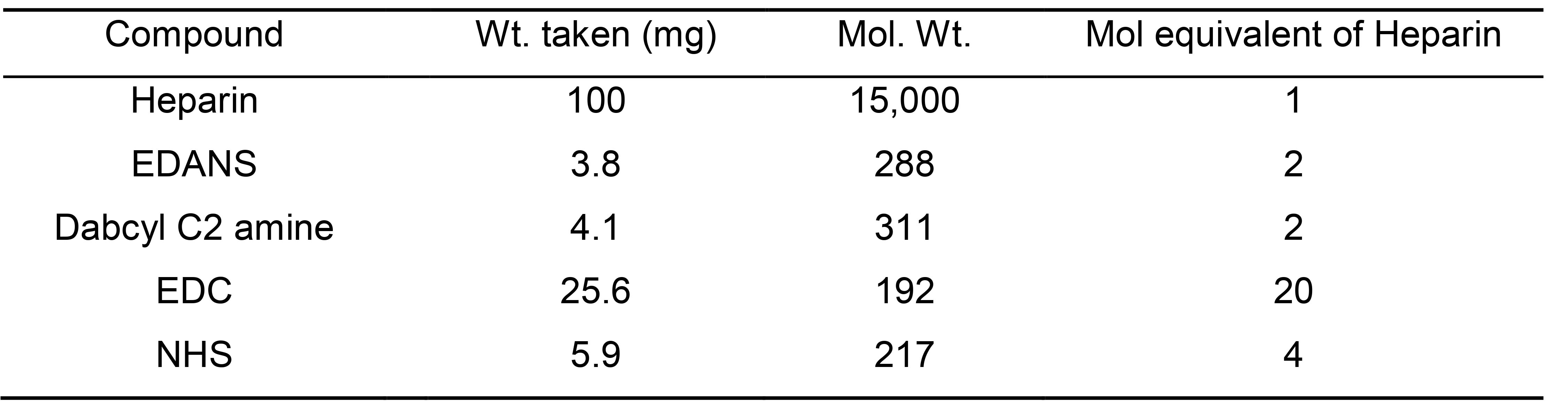
- Add DMSO dissolved mixture, from Step 4, into the vial containing heparin, NHS and EDC.
- Make up the final volume up to 2 ml with distilled H2O and drop a micro stir bar.
- Stir mixture continuously on temperature controlled magnetic stirrer.
- Continue stirring 20 h at 37 °C on an oil bath. If the oil bath is not available, a temperature-controlled oven with occasional mixing could be used as well.
Note: Make sure to stir/mix reaction mixture during labeling. - Later, dialyze mixture against distilled H2O at room temperature to remove unreacted label and salts. Use a molecular weight cut off (MWCO) of 3.5-5 kDa membrane.
Note: Dialyze in 1 L distilled H2O (dark/turn off lights) and change distilled H2O 3 times in 24 h. After dialysis, the sample volume increases from 2 to ~6/7 ml. - Next, the excess H2O and salts are removed by concentration using 3,000 Da MWCO membrane centrifugation concentrator. Repeat the centrifugation until the volume reduces to ~0.5 ml.
Note: An Eppendorf centrifuge at 4500 x g (rcf) for 20 min at 8 °C is used for this purpose. The disposable centrifugal concentrators are devices used mainly for concentration, and desalting. Depending upon the manufacturers, these are called as centricon or centrifugal filter unit. - As a final step, load concentrated sample onto the G-15 size exclusion column.
Note: This technique elutes/separates the mixture based on molecular weight. The higher molecular weight compounds elute early followed by the low molecular weight later. Since the heparin molecular weight ~15,000 Da, use Sephadex G-15 (small biomolecules greater than 1500 molecular weight) column for desalting.
The Sephadex G-15 (50 g) is washed in distilled water and allowed swell overnight. Next morning, the floating fine particles are decanted, before packing the column. Pack column (24 ml) under gravity and equilibrate overnight with water using a peristaltic pump at a flow rate of 0.5 ml/min. - Subsequently, load concentrated labeled (0.5 ml) heparin gently onto the equilibrated column using a glass dropper and elute with water at 0.5 ml/min. The colored fractions of size 1 ml are collected manually in conical tubes and absorbance is recorded at 500 nm using 1 cm path length cuvette (Figure 1).
Note: The tubes are weighed previously to calculate the product final product weight (WF - WI = Product weight).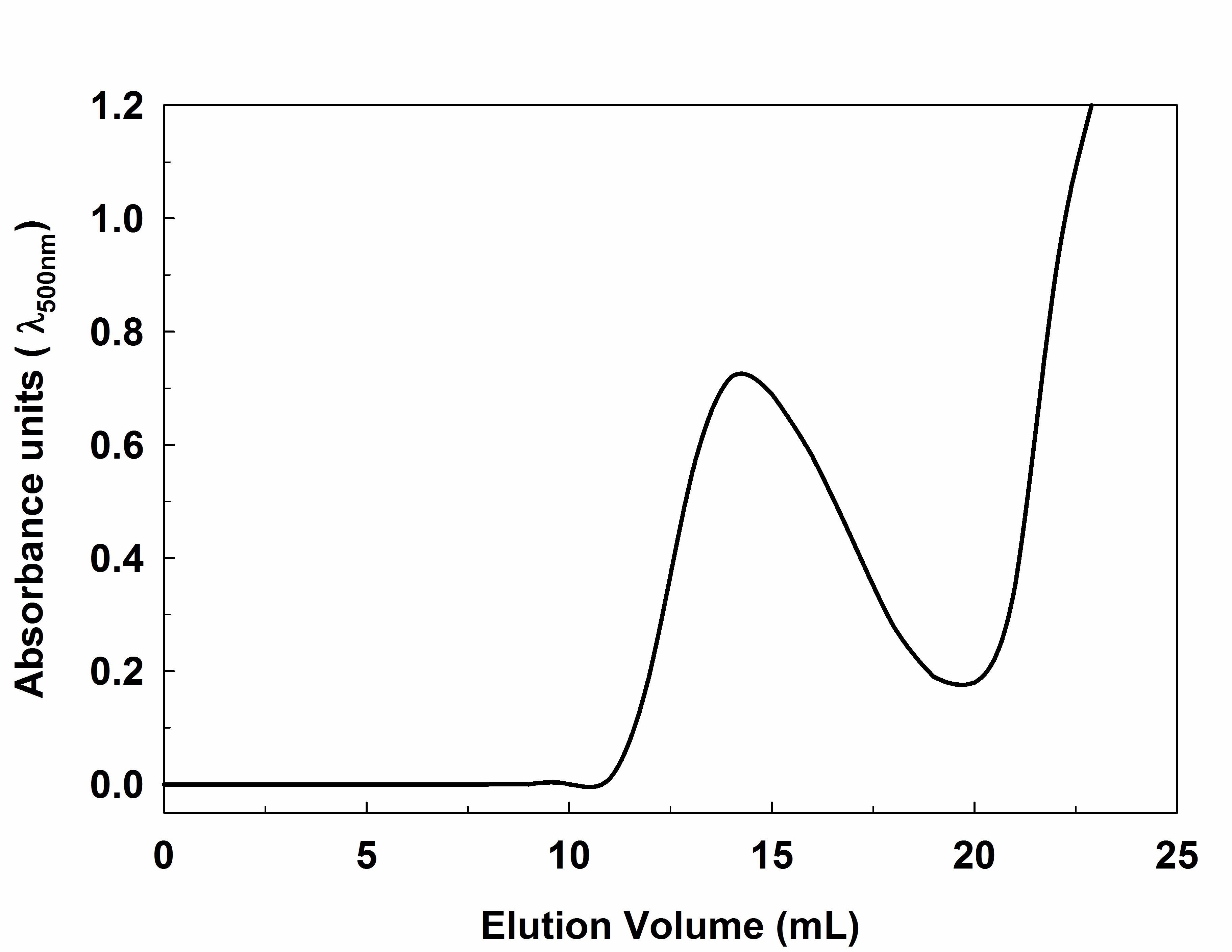
Figure 1. Labeled heparin fractionated on G-15 column. The fractions absorbance is recorded at 500 nm and plotted against the elution volume. - The absorbance spectrum of EDANS labeled, Dabcyl labeled, and Dabcyl-EDANS labeled Heparin is shown in Figure 2. In addition, Figure 3 shows the lyophilized labeled heparin, 10 mg/ml solution and the 96-well plate excited at 360 nm (1 mg/ml).
- Freeze fluorescent colored fractions in -80 °C for 3 h followed by Lyophilization. Further, labeling is confirmed using NMR (Figure 4). Prepare each (10 mg/ml) Labeled heparin, Heparin, EDANS labeled Heparin and Dabcyl labeled heparin in D2O.
Note: EDANS labeled and Dabcyl Labeled heparin are prepared similarly as explained above. Step 4 will have either EDANS (EDANS labeled heparin) or Dabcyl (Dabcyl labeled heparin). - Carefully fill the NMR tubes (500 μl) and record the spectra. Here Heparin is used as the reference.
- Further experimental details can be found on Sistla et al. (2019).
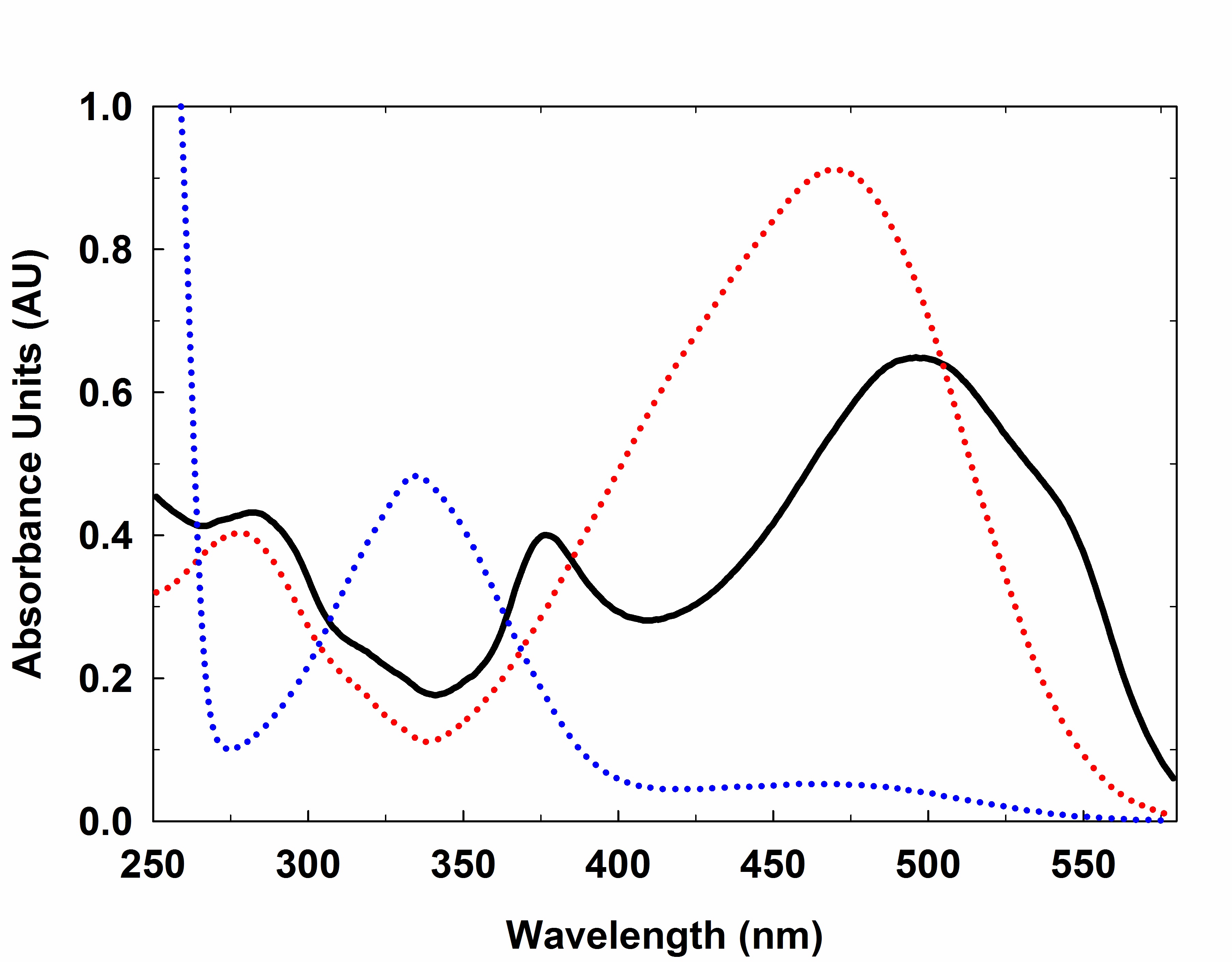
Figure 2. Spectra representative of labeled heparin. (...) (red dotted line) Heparin-Dabcyl; (...) (blue dotted line) Heparin-EDANS; and (―) (solid line) Heparin Dabcyl-EDANS. The spectra are recorded from 600-250 nm by dissolving the labeled heparin in water using 1 cm path length quartz cuvette.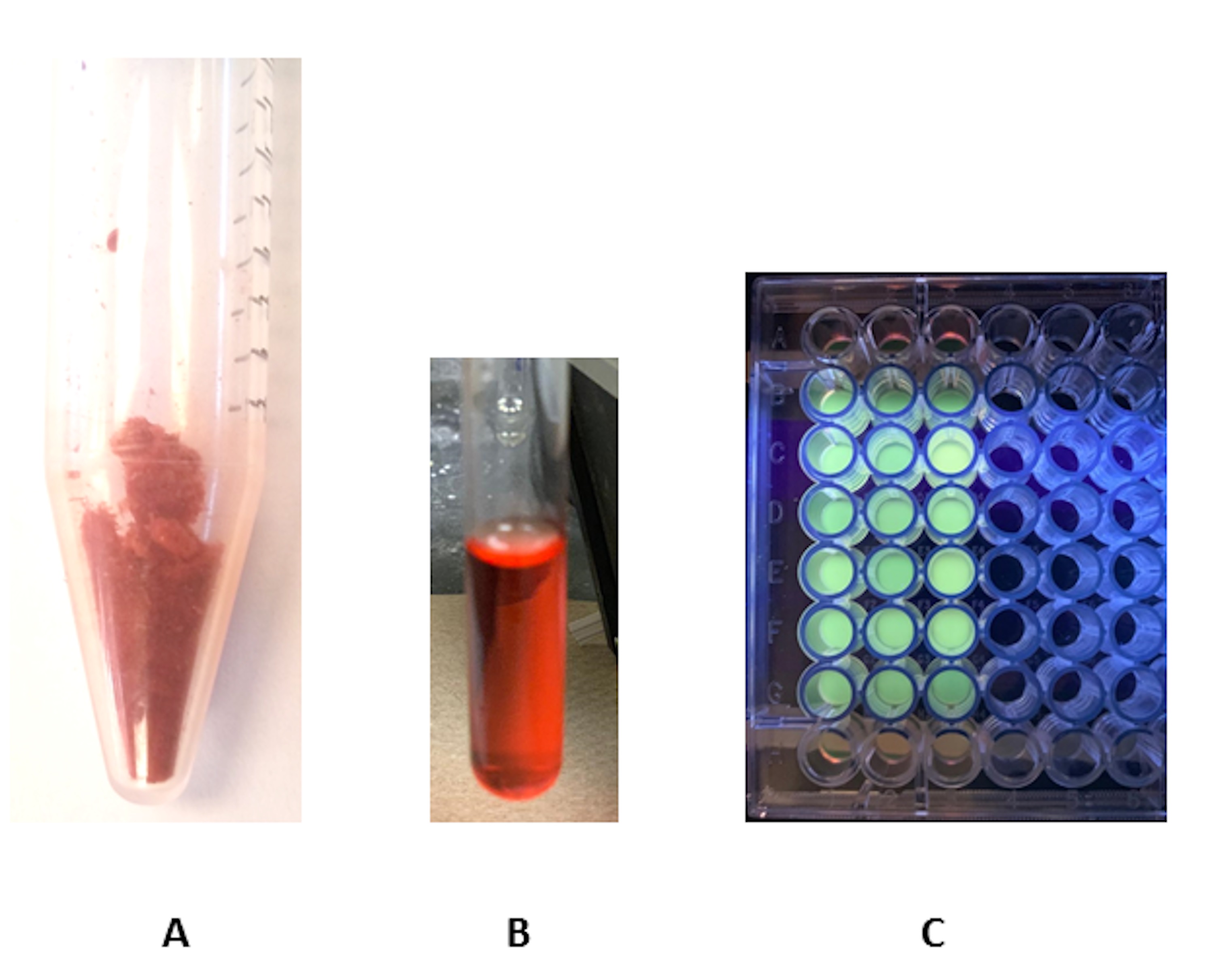
Figure 3. Sample preparation workflow. A. Labeled heparin; B. Labeled heparin (10 mg/ml) in assay buffer; C. Labeled heparin solution (1 mg/ml) in a 96-well plate showing the EDANS fluorescence when excited at 360 nm.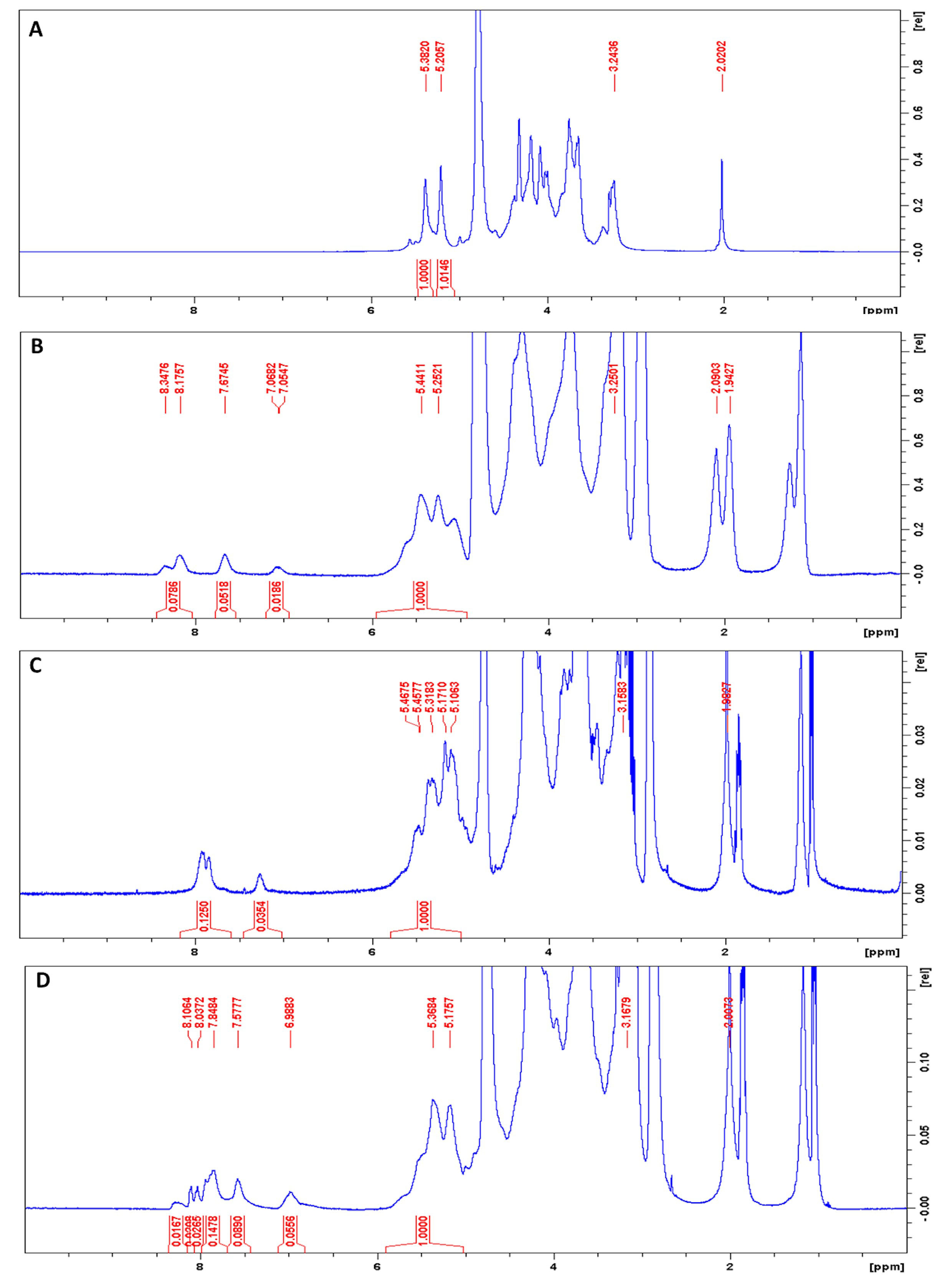
Figure 4. 400 MHz 1H NMR spectra of heparin and FRET-labeled heparins in D2O. Peaks at 5.43 and 5.26 ppm indicate the anomeric protons in heparin, whereas peaks in the range of 7-9 ppm correspond to EDANS and Dabcyl C2 amine. A. Unlabeled heparin; B. Heparin labeled with EDANS; C. Heparin labeled with Dabcyl; D. Heparin labeled with EDANS and Dabcyl.
- Sample preparation and data acquisition
- Heparanase protein expression and purification
Heparanase protein is expressed and purified using the published method (Wu et al., 2015; Sistla et al., 2019). The protein could also be obtained from R&D systems. Aliquot the stock protein and store at -80 °C until use. - Assay
- Prepare Sodium acetate buffer 0.2 M pH 5. (see Recipe 1)
- A solution containing recombinant heparanase (1 μM) and the labeled substrate (1 mg/ml) is vigorously shaken for 4 h.
- Inactivate enzyme by incubating on ice until the measurements are read.
- Transfer the mixture to a fluorescence cuvette and record the emission spectrum from 350 to 600 nm (λEX = 340 nm). The excitation and emission slits are set to 0.5 mm (Figure 5).
- Reactions are set up either in duplicate or triplicate.
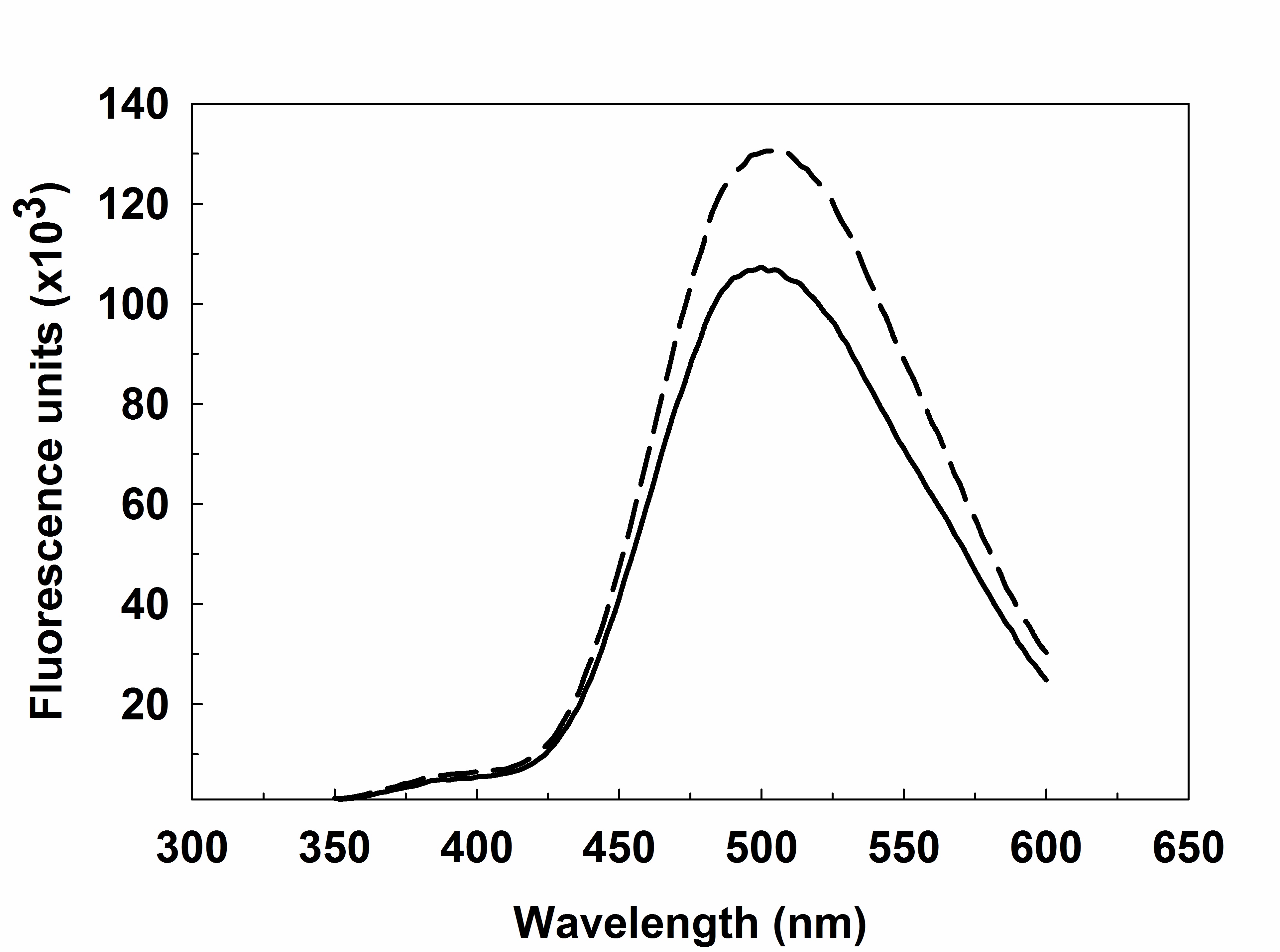
Figure 5. FRET-based assay for human heparanase. Labeled heparin (1 mg/ml) is incubated with heparanase (1 μM) at 37 °C in 20 mM sodium acetate buffer, pH 5.0. Fluorescence emission spectra (λEX = 340 nm) at (bold line) 0 h, (dotted line) 4 h.
- Heparanase protein expression and purification
- Inhibition assay–Plate format
Inhibition assay is performed using varying concentrations of Inhibitor. The hydrolysis of labeled heparin by heparanase in the presence of suramin, a known inhibitor, is monitored in 96-well plates (Corning) on a fluorescence microplate reader Flex Station III (Molecular Devices) in a final volume of 100 μl.- Prepare a stock solution of the inhibitor in the assay buffer.
- Incubate heparanase (1 μM) and various concentrations of suramin for 30 min in the assay buffer at 37 °C.
- Initiate the hydrolytic reaction by the addition of labeled heparin (66 μM final concentration).
- Stir constantly for 4 h, and terminate the reaction by placing on ice.
- Record fluorescence reading at 500 nm (λEX = 340 nm, Figure 6).
- The IC50 is calculated using the standard method. The residual activity is calculated comparing to control with no inhibitor.
- Plot a semi-logarithmic graph, concentration versus residual activity.
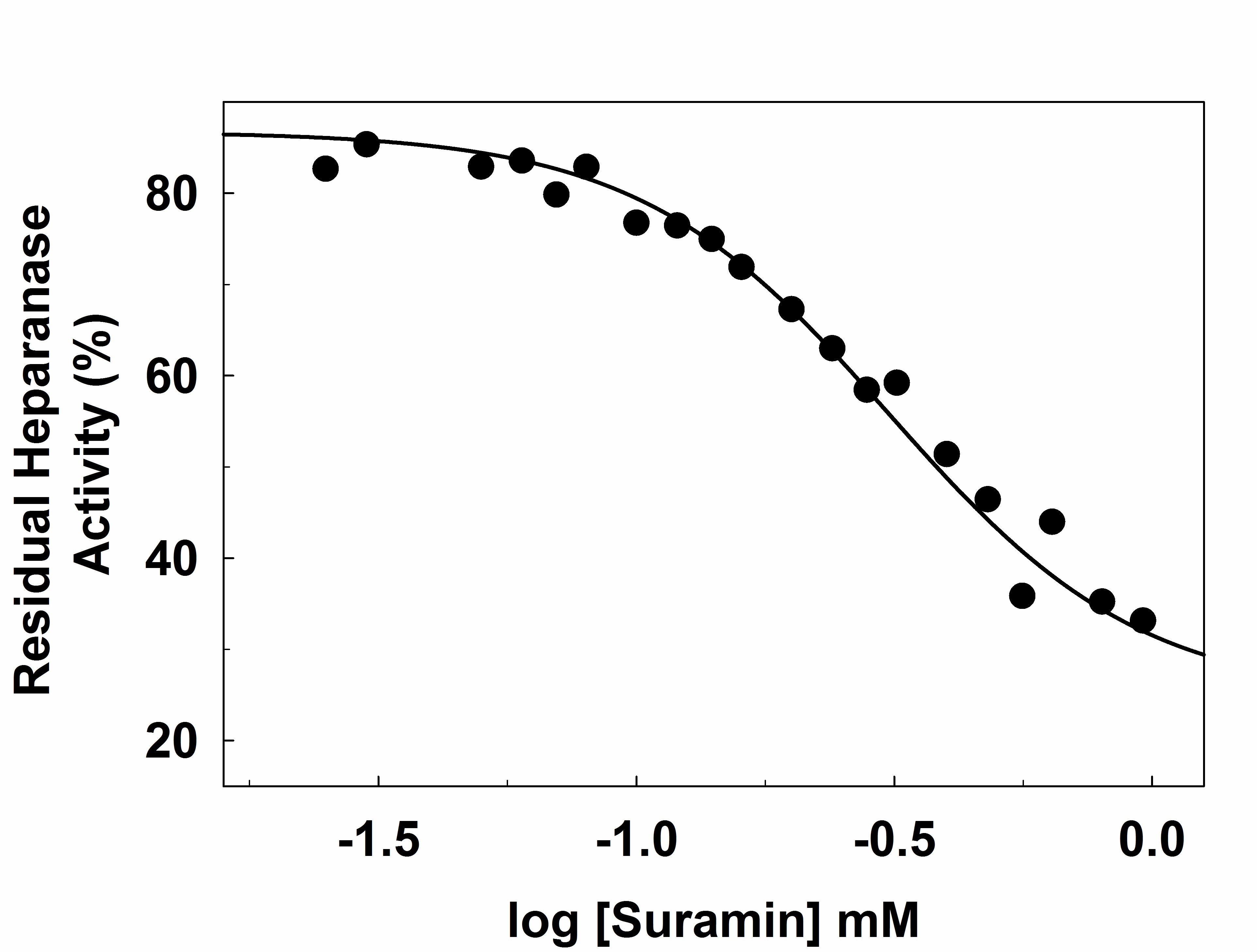
Figure 6. Suramin inhibition of heparanase. The experiment is performed in microplate format (100 μl) in 20 mM sodium acetate buffer, pH 5.0, containing 1 mg/ml labeled heparin and 1 μM heparanase at 37 °C for 4 h in the presence of varying concentrations of suramin. Solid lines represent curve fitting to standard dose-response equation.
- Detection heparanase activity in biological samples
- The human mammary carcinoma cell line, MCF7 and HEK 293T (negative control) cell lines are maintained in monolayer conditions in Dulbecco's Modified Eagle Medium (DMEM) growth media (Invitrogen, USA), supplemented with 10% fetal bovine serum (FBS), and 1% antibiotic-antimycotic liquid (AA) in 5% CO2 at 37 °C, as recommended by ATCC.
- Initially, grow MCF7/HEK cells as a monolayer in the growth media for 24 h to ensure efficient growth and adhesion.
- Subsequently, replace the media with 2% FBS serum in growth media and allow the cells to propagate for an additional 48 h in normoxia conditions (humidified atmosphere maintained at 37 °C in 5% CO2/95% atmospheric air).
- After 48 h of incubation, the supernatants are collected and centrifuged at 1,000 x g for 10 min at 4 °C.
- For the time-based heparanase activity measurements, harvest media at varying time points and freeze immediately at -20 °C until the assay is performed.
- Media containing heparanase is added to labeled heparin substrate to a final volume of 100 μl in 96-well plates (Corning® 3897).
- Stir constantly for 4 h, and terminate the reaction by placing on ice.
- Record fluorescence reading at 500 nm (λEX = 340 nm). The experimental results are shown in Figure 7.
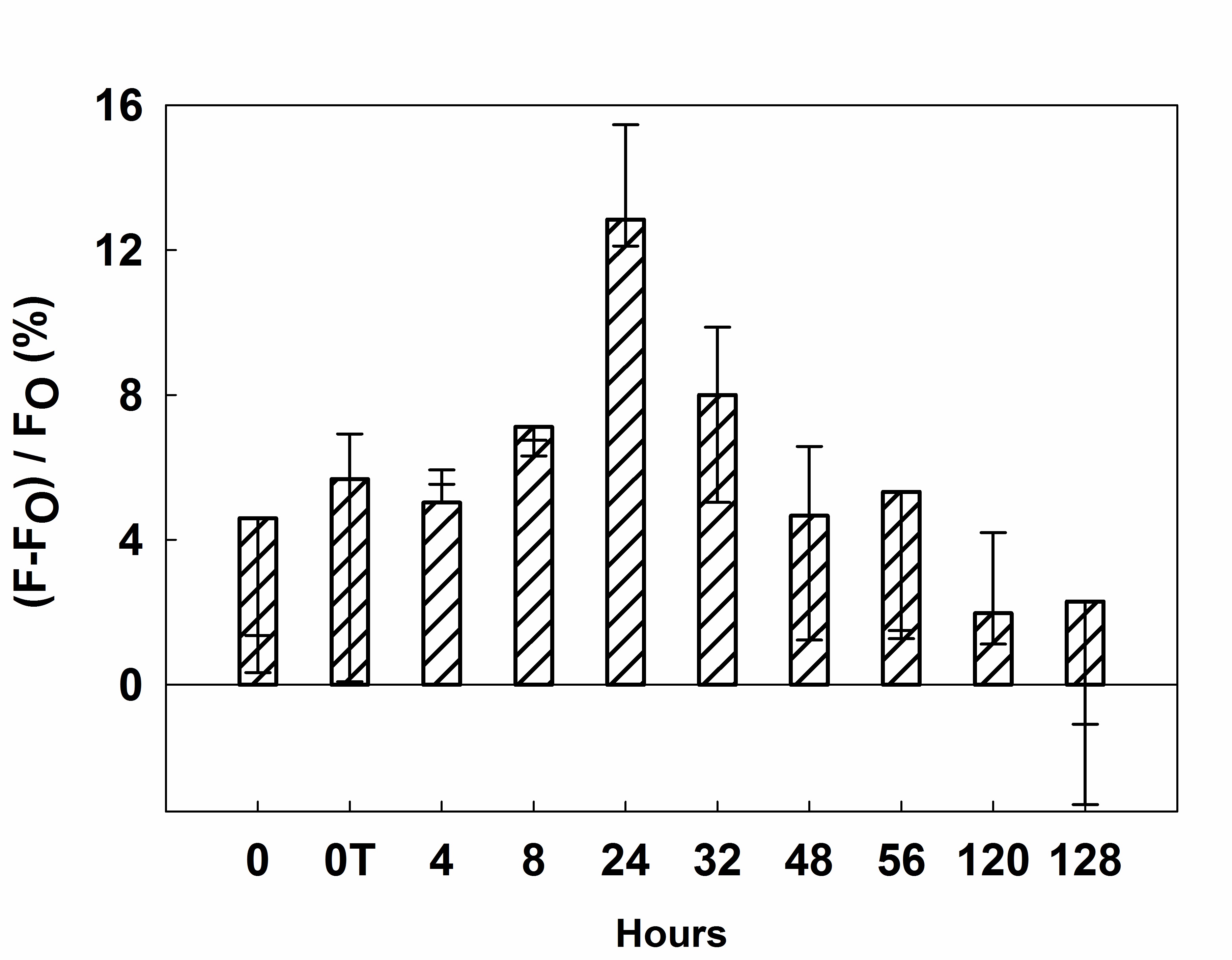
Figure 7. Expression of active heparanase by MCF7 cells under normoxic conditions. Results are presented as the mean ± SD (n > 3).
Data analysis
The observed change in fluorescence (ΔF) is normalized to initial fluorescence (Fo) to calculate the % change in signal at every addition of a test sample. The data is fit to the Standard Curves: Four Parameter Logistic Curve using Sigma Plot (IC50) / Logistic fit if OrignPro or Kaleidagraph is used.
Notes
- Use Assay buffer for the sample preparations and dilutions.
- Always prepare labeled substrate fresh in an amber centrifuge tube just before the experiment.
- Protein is kept on ice until the experiment is performed.
- The substrate has to be shaken vigorously in the dark.
- Use appropriate blanks.
- If required, dilute sample before the fluorescence measurement.
Recipes
- Assay buffer
20 mM sodium acetate buffer, pH 5.0, containing 100 mM NaCl
Labeled heparin stock: 10 mg/ml prepared fresh in assay buffer
Heparanase enzyme: 25 μM stock
Water bath: At temperature 37 °C
Reaction volume: 100 μl
Note: The enzyme assays are conducted in low retention micro centrifuge tubes.
Acknowledgments
This work was supported by grants from NIH HL107152 and HL128639 to URD.
Competing interests
The authors declare no competing financial interest.
References
- Behzad, F. and Brenchley, P. E. (2003). A multiwell format assay for heparanase. Anal Biochem 320(2): 207-213.
- Dempsey, L. A., Brunn, G. J. and Platt, J. L. (2000). Heparanase, a potential regulator of cell-matrix interactions. Trends Biochem Sci 25(8): 349-351.
- Enomoto, K., Okamoto, H., Numata, Y. and Takemoto, H. (2006). A simple and rapid assay for heparanase activity using homogeneous time-resolved fluorescence. J Pharm Biomed Anal 41(3): 912–917.
- Fairbanks, M. B., Mildner, A. M., Leone, J. W., Cavey, G. S., Mathews, W. R., Drong, R. F., Slightom, J. L., Bienkowski, M. J., Smith, C. W., Bannow, C. A. and Heinrikson, R. L. (1999). Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem 274(42): 29587-29590.
- Freeman, C. and Parish, C. R. (1997). A rapid quantitative assay for the detection of mammalian heparanase activity. Biochem J 325 (Pt 1): 229-237.
- Hammond, E., Li, C. P. and Ferro, V. (2010). Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Anal Biochem 396(1): 112-116.
- Kussie, P. H., Hulmes, J. D., Ludwig, D. L., Patel, S., Navarro, E. C., Seddon, A. P., Giorgio, N. A. and Bohlen, P. (1999). Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun 261(1): 183-187.
- Melo, C. M., Tersariol, I. L., Nader, H. B., Pinhal, M. A. and Lima, M. A. (2015). Development of new methods for determining the heparanase enzymatic activity. Carbohydr Res 412: 66-70.
- Peterson, S. B. and Liu, J. (2013). Multi-faceted substrate specificity of heparanase. Matrix Biol 32(5): 223-227.
- Sanderson, R. D., Elkin, M., Rapraeger, A. C., Ilan, N. and Vlodavsky, I. (2017). Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J 284(1): 42-55.
- Schiemann, S., Lühn, S. and Alban, S. (2012). Development of both colorimetric and fluorescence heparinase activity assays using fondaparinux as substrate. Anal Biochem 427 (1): 82–90.
- Sistla, J. C., Morla, S., Alabbas, A. B., Kalathur, R. C., Sharon, C., Patel, B. B. and Desai, U. R. (2019). Polymeric fluorescent heparin as one-step FRET substrate of human heparanase. Carbohydr Polym 205: 385-391.
- Toyoshima, M. and Nakajima, M. (1999). Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem 274(34): 24153-24160.
- Tsuchida, S., Podyma-Inoue, K. A. and Yanagishita, M. (2004). Ultrafiltration-based assay for heparanase activity. Anal Biochem 331(1): 147-152.
- Wu, L., Viola, C. M., Brzozowski, A. M. and Davies, G. J. (2015). Structural characterization of human heparanase reveals insights into substrate recognition. Nat Struct Mol Biol 22(12): 1016-1022.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sistla, J. and Desai, U. R. (2019). A Robust, One-step FRET Assay for Human Heparanase. Bio-protocol 9(17): e3356. DOI: 10.21769/BioProtoc.3356.
Category
Cancer Biology > General technique > Biochemical assays
Biochemistry > Carbohydrate > Oligosaccharide > Labelling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









