- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Differentiation of Thymic Treg Cell Progenitors to Mature Thymic Treg Cells
Published: Vol 9, Iss 16, Aug 20, 2019 DOI: 10.21769/BioProtoc.3335 Views: 4706
Reviewed by: Alka MehraChris TibbittAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Differentiation, Maintenance, and Contraction Profiling of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes
Matthijs Snelders [...] Jeroen Essers
Mar 5, 2025 3966 Views

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2432 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 269 Views
Abstract
Thymic Treg cell differentiation occurs via a two-step process. Step one generates Treg cell progenitors (TregP) and is driven by strong TCR interactions with antigens presented in the thymus. Step two is initiated by activation of STAT5 via IL-2, or IL-15, leading to mature Treg cells capable of emigrating from the thymus and mediating immune tolerance and homeostasis in peripheral tissues. Herein we describe an in vitro TregP cell differentiation assay that models the second, cytokine dependent, step of thymic Treg cell development. It can be utilized with relative ease to determine if a population of thymocytes represents a potential progenitor population for Treg cells as well as test how different cytokines or chemical inhibitors modulate the differentiation of known TregP cell populations into mature Treg cells.
Keywords: ThymusBackground
Regulatory T cells (Treg) cells are a population of T cells that can suppress immune responses and are required to maintain immune tolerance and tissue homeostasis. While two broad classes of Treg cells have been described, thymic Treg cells (tTreg) and peripheral Treg cells (pTreg), the majority of Treg cells are generated during thymic selection (Asano et al., 1996). Early experiments describing autoimmune manifestations in thymectomized mice, which can be rescued by Treg cell transfer, confirm the importance of the thymus in generating Treg cells capable of maintaining immune homeostasis (Asano et al., 1996). The development of Treg cells in the thymus is a two-step process (Burchill et al., 2008; Lio and Hsieh, 2008). The first step is TCR mediated and gives rise to CD25+Foxp3- or CD25-Foxp3lo TregP cells. The second step is mediated by contact of these progenitor populations with intrathymic cytokines leading to co-expression of CD25 and Foxp3 and generation of mature tTreg cells.
The in vitro TregP conversion assay models the second, cytokine dependent, step in tTreg cell development. This assay builds on protocols initially described by work from Chyi Hsieh’s lab and modified by our own lab (Burchill et al., 2008; Lio and Hsieh, 2008). The technique described below provides a simple method to determine if an isolated thymic population represents a TregP, characterized by independence from TCR stimulation and upregulation of tTreg markers in response to cytokine. This assay has been performed with varied stimulation conditions previously, including stimulation with high concentrations of IL-2 (200 U/ml) (Tai et al., 2013) as well as IL-2 + IL-7 (Lio and Hsieh, 2008). The protocol we describe converts bona-fide TregP to mature Treg cells robustly and reproducibly with small amounts of IL-2 (Mahmud et al., 2014; Owen et al., 2019). As such, investigators can narrow the application of IL-2 to concentrations that are more physiologically relevant. This assay has also been able to detect conversion of TregP to mature Treg cells with other cytokines like IL-7, IL-15 and IL-4, as well as combination of factors such as IL-2 with TNFRSF agonists (Mahmud et al., 2014). Thus, the in vitro TregP conversion assay can be used to query various important stimulatory, or inhibitory, signals that may be encountered in vivo by developing TregP.
Materials and Reagents
- Pipette tips
- 0.22 μm 1,000 ml Stericup (Millipore Sigma, catalog number: SCGPU11RE)
- 0.22 μm 50 ml Steriflip (Millipore Sigma, catalog number: SCGP00525)
- LS Columns (Miltenyi Biotec, catalog number: 130-042-101)
- 50 ml Falcon conical tube (Corning, catalog number: 352070)
- 15 ml CentriStar conical tube (Corning, catalog number: 430790)
- 5 ml Falcon Round Bottom test tube (Corning, catalog number: 352052)
- 96-well round bottom, non-TC treated plates (Sarstedt, catalog number: 82.1582.001)
- CytoOne Non-treated 6-well plates (USA Scientific, catalog number: CC7672-7506)
- Falcon 70 μm cell stainer (Corning, catalog number: 352350)
- Plain and frosted glass slides (VWR International, catalog number: 48312-004)
- Foxp3GFP (B6.Cg-Foxp3tm2(EGFP)Tch/J) mice (THE JACKSON LABORATORY, catalog number: 006772)
The best TregP cell yields come from younger (5-6 weeks old) female mice due to a generally large thymus. If younger mice are not available older mice will provide usable TregP cells. However, the number of TregP cells recovered could be reduced by 50% of that obtained from younger mice. - Recombinant Human IL-2 (R&D Systems, catalog number: 202-IL)
- Anti-mouse CD4 (GK1.5) BV786 (1:200) (BD Biosciences, catalog number: 563331)
- Anti-mouse CD8a (53-6.7) Biotin (1:200) (Tonbo Biosciences, catalog number: 30-0081-U500)
- Anti-mouse CD25 (PC61.5) PerCP-Cy5.5 (1:200) (Tonbo Biosciences, catalog number: 65-0251-U100)
- Anti-mouse CD73 (eBioTY/11.8) eF450 (1:100) (eBioscience, catalog number: 48-0731-82)
- Anti-mouse Ter119 (Ter-119) Biotin (1:200) (BD Biosciences, catalog number: 553672)
- (Optional) antibodies:
- Anti-CD11b (M1/70) Biotin (1:200) (Tonbo Biosciences, catalog number: 30-0112-U500)
- Anti-CD11c (N418) Biotin (1:200) (Tonbo Biosciences, catalog number: 30-0114-U500)
- Anti-NK1.1 (PK136) Biotin (1:200) (Tonbo Biosciences, catalog number: 30-5941-U500)
- Anti-γδTCR (eBioGL3) Biotin (1:200) (eBioscience, catalog number: 13-5711-85)
- Anti-B220 (RA3-6B2) Biotin (1:200) (Tonbo Biosciences, catalog number: 30-0452-U500)
- Streptavidin APC-eF780 (1:200) (eBioscience, catalog number: 47-4317-82)
- Viability Dye GhostTM Red 780 (1:1,000) (Tonbo Biosciences, catalog number: 13-0865-T100)
- Streptavidin Microbeads (Miltenyi Biotec, catalog number: 130-048-101)
- AccuCheck Counting Beads (Life Technologies, catalog number: PCB100)
- Sodium azide (NaN3) (Ricca Chemical, catalog number: 7144.8-16)
- 10x PBS (Fisher BioReagents, catalog number: BP399-20)
- Ethylenediaminetetraacetic Acid (EDTA) (Fisher Scientific, catalog number: BP120-500)
- Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11150)
- RPMI 1640 media (Corning, catalog number: 15-040-CV)
- Penicillin-Streptomycin Solution (Corning, catalog number: 30-002-CL)
- L-Glutamine (Corning, catalog number: 25-005-CL)
- MEM Nonessential amino acids (Corning, catalog number: 25-025-CL)
- 2-mercaptoethanol (MP Bio, catalog number: 02194705-CF)
- HEPES (Corning, catalog number: 25-060-CL)
- Sodium Pyruvate (Corning, catalog number: 25-000-CL)
- Sort Buffer (see Recipes)
- FACS Buffer (see Recipes)
- cRPMI (see Recipes)
Equipment
- Pipettes
- Sorvall Legend X1R Centrifuge (Thermo Scientific, catalog number: 75004260)
- BD Biosciences LSR II (BD, model: LSR II)
- BD Biosciences FACSAria II (BD, model: FACSAria II)
- MACS Multistand (Miltenyi Biotec, catalog number: 130-042-303)
- QuadroMACS Separator Magnet (Miltenyi, catalog number: 130-090-976)
- Class II Biosafety Cabinet/Laminar Flow Hood
- IncuSafe Incubator (Panasonic, catalog number: KM-CC17T0A)
- Hemocytometer (Fisher, catalog number: 0267110)
- Refrigerator (2-8 °C, Kenmore)
- Panasonic -80 °C VIP Plus Freezer
Software
- FlowJo 10 (10.5.3) (https://www.flowjo.com/)
Procedure
- TregP cell isolation
- Euthanize mice by CO2 inhalation.
- Harvest thymi from mice into 5 ml Sort buffer in 6-well plates. The number of conditions tested and replicates determine the number of mice required for each experiment. In a simple experiment, 1 mouse can be used as this should provide 5-10 x 104 CD25+ and Foxp3lo TregP cells.
- Work in a clean biosafety cabinet from this point on.
- Mechanically dissociate thymi between frosted glass slides or favored method of mechanical dissociation.
- Transfer cells into 50 ml conical vial through 70 μm filter.
- Spin cells at 350 x g for 10 min, at 4 °C (keep thymi in individual 50 ml conical tubes).
- Resuspend cells in 1 ml depletion cocktail of biotin-labeled antibodies. This cocktail of antibodies should include anti-CD8 (1:200) and anti-Ter119 (1:200). However other optional depletion antibodies can be added to this cocktail such as anti-CD11b (1:200), anti-CD11c (1:200), anti-NK1.1 (1:200), anti-γδTCR (1:200) and anti-B220 (1:200).
- Incubate on ice for 20 min and cover to protect from light.
- Add 15 ml Sort buffer; spin cells at 350 x g for 5 min, at 4 °C.
- Count the cells and resuspend in Strepavidin microbead cocktail (~1 μl beads per 2-3 million cells diluted in Sort buffer to a final volume of ~400 μl/thymus).
- Incubate for 20 min at 4 °C (agitate by swirling tube gently after 10 min).
- Add 10 ml Sort buffer; spin cells at 350 x g for 5 min, at 4 °C.
- Pre-rinse LS columns (1 column/thymus) with 3 ml cold Sort Buffer.
- Resuspend cells in 2 ml Sort buffer.
- Apply cells from individual thymus to separate LS columns on QuadroMACS Miltenyi separator magnet through a filter (Figure 1).
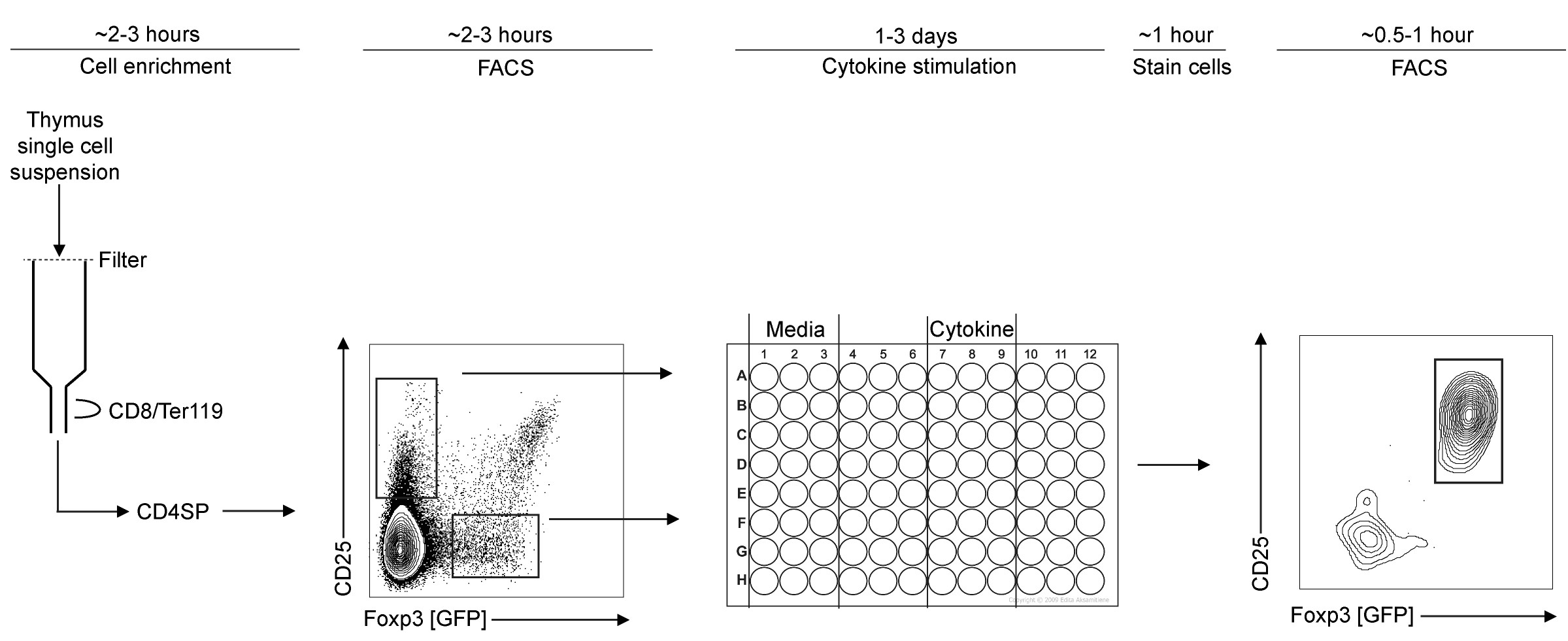
Figure 1. Schematic of in vitro TregP differentiation assay workflow. First, thymi are processed and enriched for CD4 single positive (CD4SP) thymocytes. This fraction is stained for surface markers then each TregP is isolated by FACS. Isolated TregP are stimulated with cytokine in 96-well plate for a pre-determined time. These cells are then harvested and analyzed by flow cytometry for upregulation of Foxp3 and/or CD25. - Wash columns 3 times each with 3 ml cold Sort buffer.
- Collect unbound fraction in 15 ml conical vials; spin cells at 350 x g for 5 min, at 4 °C.
- Combine cells into a single 15 ml conical tube; apply surface staining cocktail [CD4 (1:200), Live-Dead (1:1,000), CD25 (1:200), CD73 (1:100) and Streptavidin-APC-eF780 (1:200)]. The Streptavidin-APC-eF780 is used to detect biotin-conjugated antibodies used for magnetic depletion, including CD8, Ter119 and any other optional antibodies included from Step 7 above.
- Incubate on ice for 20 min and cover to protect from light.
- Add 14 ml Sort buffer; spin cells at 350 x g for 5 min, at 4 °C.
- Resuspend cells in 1 ml Sort buffer and filter cells through a 70 μm filter into a new, pre-chilled 15 ml conical; wash the filter with Sort buffer to bring cells to appropriate final volume.
- Set up gates on FACS Aria II for sorting TregP cells as described in Figure 2.
- Collect CD25+ and/or Foxp3lo TregP cells into separate 5 ml round bottom tubes with 1.5 ml Catch buffer (1x PBS + 50% FBS).
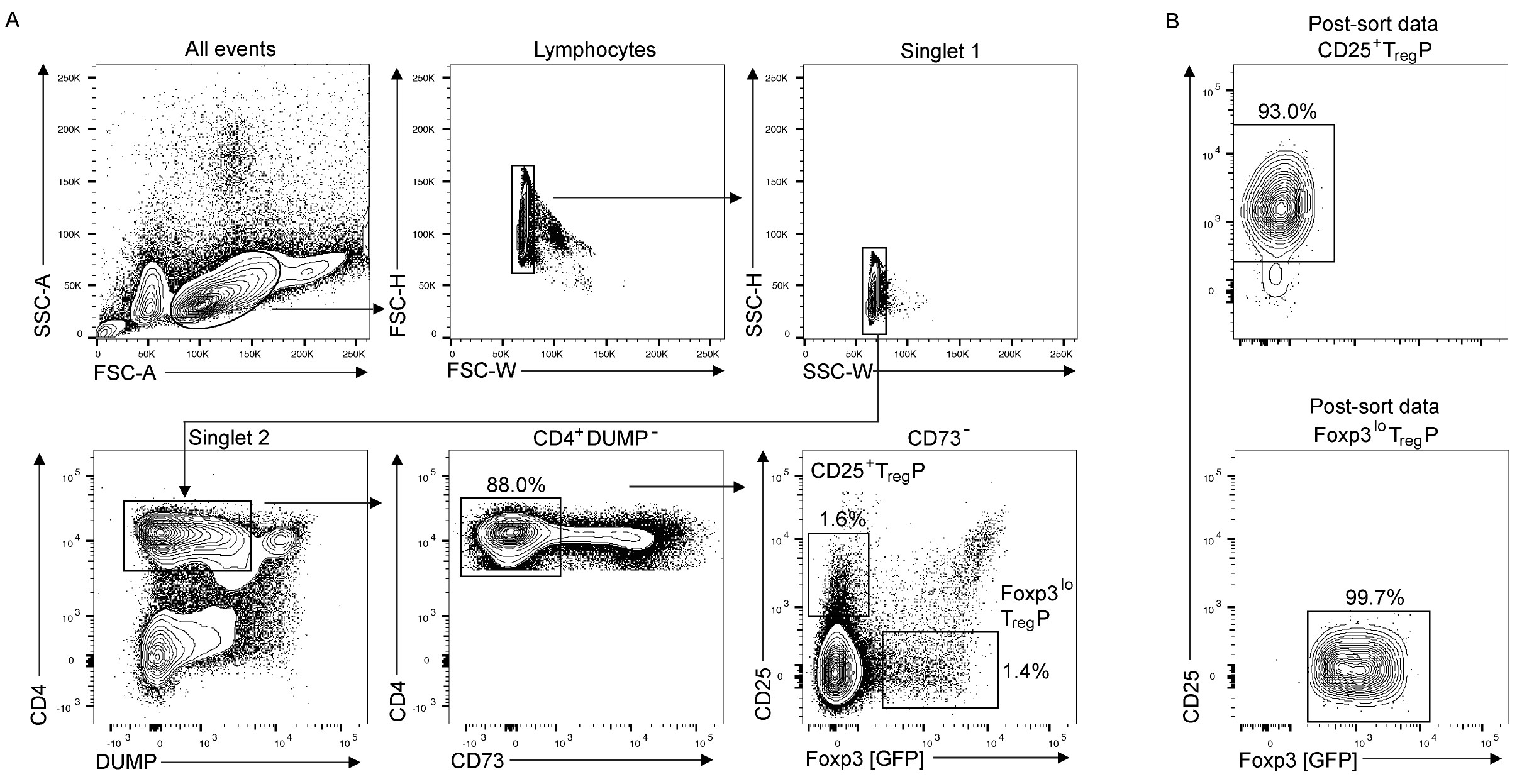
Figure 2. Flow cytometry gating strategy for sorting TregP. A. Flow cytometry gating scheme for sorting TregP from CD8/Ter119 depleted thymocytes. DUMP channel contains CD8, Ter119 (these are labeled with biotin conjugated antibodies during magnetic depletion that are probed with APC-eF780-conjugated streptavidin) and Live-Dead. CD73+ cells represent recirculating and long-term resident TregP and Treg. Thus sorting CD73- thymocytes ensures analysis of de novo developing TregP. B. Post-sort purity analysis of CD25+ TregP (top) and Foxp3lo TregP (bottom).
- Stimulation of TregP cells
- Make stimulation media (example: 2 U/ml IL-2 in cRPMI for a final concentration of 1 U/ml).
- Plate 100 μl of stimulation media into each well of non-TC treated round bottom 96-well plate.
- Place the 96-well plate in 37 °C incubator.
- Fill 5 ml round bottom tubes containing sorted TregP cells with cRPMI, spin cells at 350 x g for 5 min, at 4 °C.
- Resuspend cells at 1-2 x 105 cells/ml in cRPMI.
- Add 100 μl of cells to 100 μl stimulation media in 96-well plates (plate 1-2 x 104 cells/well).
- Place the 96-well plates into 37 °C incubator.
- Incubate for 24-72 h (we perform these assays for 72 h typically but CD25 and/or Foxp3 upregulation can be observed at earlier timepoints).
- Analysis of in vitro TregP differentiation assay
- Add 100 μl FACS buffer to each well of stimulated TregP.
- Spin cells at 350 x g for 5 min at 4 °C.
- Decant supernatant and add 200 μl FACS buffer.
- Spin cells at 350 x g for 5 min at 4 °C.
- Resuspend cells in 50 μl staining cocktail [anti-CD4 (1:200), anti-CD8 (1:200), anti-CD25 (1:200), Live-Dead (1:1,000)].
- Incubate on ice for 20 mins and cover to protect from light.
- Add 200 μl FACS buffer.
- Spin cells at 350 x g for 5 min at 4 °C.
- Resuspend each sample in FACS buffer/counting bead master mix (190 μl FACS buffer + 10 μl counting beads/sample).
- Analyze differentiation of TregP to mature Treg cells (typically co-expression of CD25 and Foxp3).
Data analysis
- Data analysis was performed in FlowJo 10.
- Gating scheme for quantification of Treg differentiation is shown in Figure 3. Briefly, lymphocytes were gated followed by singlets. CD4+, CD8/Live-Dead- singlets were gated and displayed for expression of CD25 and Foxp3. Representative data can be found in Owen et al. (2019) in Figure 7A, Supplemental Figure 1 and Supplemental Figure 7.
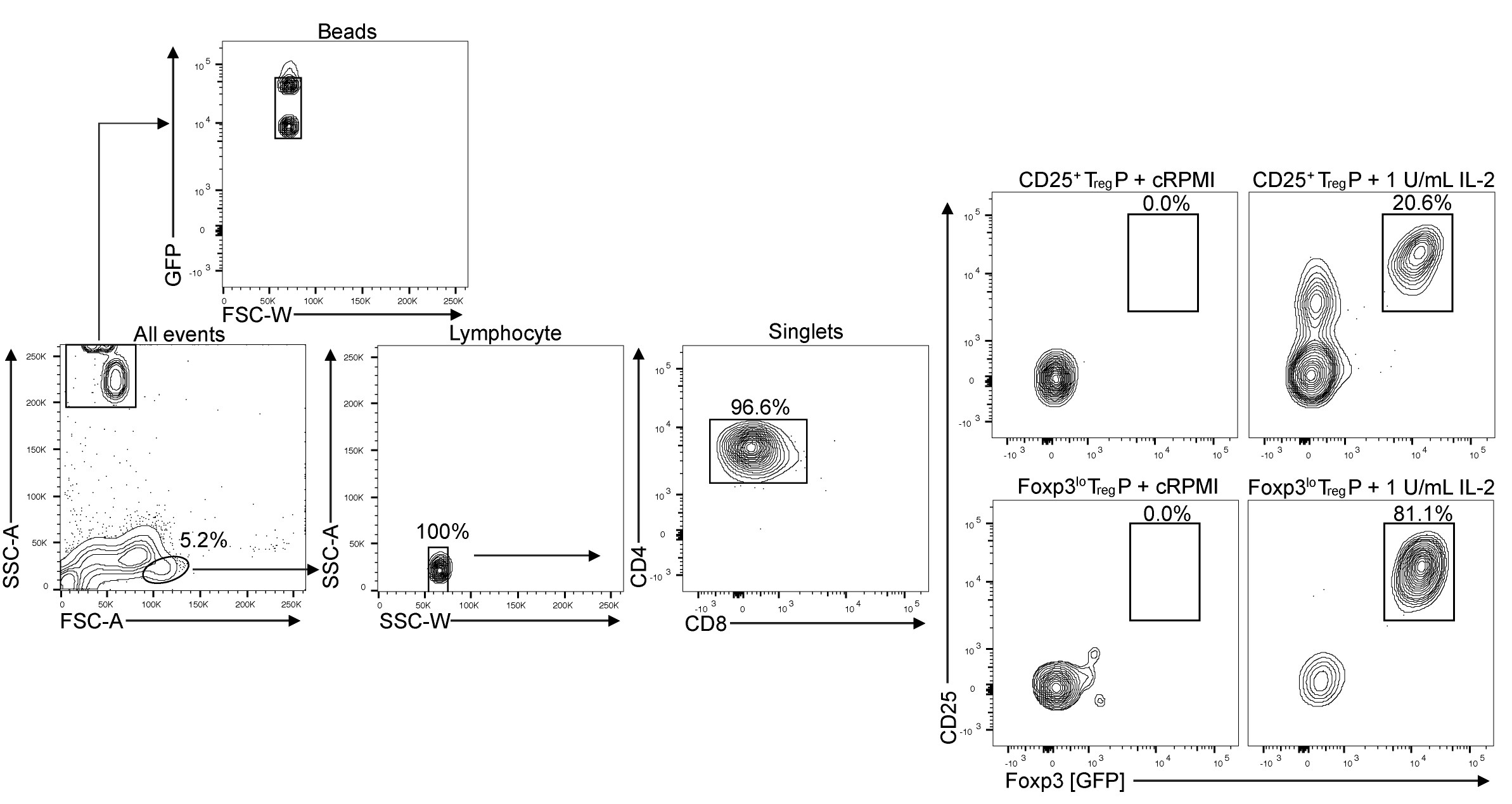
Figure 3. Flow cytometry gating scheme for analysis of TregP following 3 days of stimulation. Media alone (left) fails to convert TregP to Treg cells while 1 U/ml IL-2 stimulation (right) converts TregP to Treg cells. Beads are used to calculate total numbers of Treg cells generated in each sample. The formula used is Total cells = Cells detected x (Known Bead number added/Beads detected). - Published results (Burchill et al., 2008; Lio and Hsieh, 2008; Vang et al., 2008; Mahmud et al., 2014; Owen et al., 2019) have indicated successful conversion of CD25+ or Foxp3lo TregP is represented by dual expression of CD25 and Foxp3 in response to IL-2. However, other cytokines we have tested, such as IL-4 (Owen et al., 2019), have led to generation of CD25+ Foxp3+ Treg cells and the maintenance of CD25-Foxp3lo cells. Interestingly, IL4 stimulation of CD25-Foxp3lo TregP cells results in the development of CD25+Foxp3+ Treg cells that express distinctly lower levels of CD25 than observed with IL-2 stimulation. Thus, depending on the condition, it is relevant to also consider the proportion of CD25-Foxp3+ cells as well as the mean fluorescence intensity of CD25 and Foxp3, as shown in Owen et al. (2019) Supplemental Figure 7.
Notes
- In our hands, TregP cells are a delicate population and quite susceptible to cell death. Given this, gentle processing throughout the cell isolation protocol will yield higher amounts of TregP cells from thymi.
- A typical yield from 1 thymus is ~50,000-100,000 TregP depending on age (5-6 weeks being ideal) and sex (female having larger thymi).
- We make fresh cRPMI for each assay to help maintain viability of TregP cells in vitro.
- Our greatest success has been using round bottom, non-TC treated 96-well plates. Other 96-well formats led to increased cell death in our hands.
Recipes
- Sort Buffer (store at 4 °C, ~55-60 ml/sample)
1x PBS
2% FBS
2 mM EDTA - FACS Buffer (store at 4 °C, ~1 ml/well)
1x PBS
2% FBS
2 mM EDTA
0.05% NaN3 - cRPMI (filter sterilize, store at 4 °C)
RPMI 1640 Medium
10% Fetal Bovine Serum
1% Penicillin/Streptomycin
1% L-Glutamine
1% Sodium Pyruvate
1% Non-essential amino acids
10 mM HEPES
50 μM 2-mercaptoethanol
Acknowledgments
This work was supported by R01AI124512 (MAF) and an Immunology training grant 2T32AI007313 (DLO) from the NIH. This protocol is adapted from methods originally published by Lio and Hsieh (2008) and modified for studies from our lab (Burchill et al., 2008; Vang et al., 2008; Mahmud et al., 2014; Owen et al., 2019).
Competing interests
The authors declare no competing financial or non-financial interests.
Ethics
All animal experimentation in this protocol was approved by the University of Minnesota Institutional Animal Care and Use Committee (most recent IACUC protocol 1904-36975A, valid 5/16/2019-5/15/2022).
References
- Asano, M., Toda, M., Sakaguchi, N. and Sakaguchi, S. (1996). Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med 184(2): 387-396.
- Burchill, M. A., Yang, J., Vang, K. B., Moon, J. J., Chu, H. H., Lio, C. W., Vegoe, A. L., Hsieh, C. S., Jenkins, M. K. and Farrar, M. A. (2008). Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28(1): 112-121.
- Lio, C. W. and Hsieh, C. S. (2008). A two-step process for thymic regulatory T cell development. Immunity 28(1): 100-111.
- Mahmud, S. A., Manlove, L. S., Schmitz, H. M., Xing, Y., Wang, Y., Owen, D. L., Schenkel, J. M., Boomer, J. S., Green, J. M., Yagita, H., Chi, H., Hogquist, K. A. and Farrar, M. A. (2014). Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat Immunol 15(5): 473-481.
- Owen, D. L., Mahmud, S. A., Sjaastad, L. E., Williams, J. B., Spanier, J. A., Simeonov, D. R., Ruscher, R., Huang, W., Proekt, I., Miller, C. N., Hekim, C., Jeschke, J. C., Aggarwal, P., Broeckel, U., LaRue, R. S., Henzler, C. M., Alegre, M. L., Anderson, M. S., August, A., Marson, A., Zheng, Y., Williams, C. B. and Farrar, M. A. (2019). Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol 20(2): 195-205.
- Tai, X., Erman, B., Alag, A., Mu, J., Kimura, M., Katz, G., Guinter, T., McCaughtry, T., Etzensperger, R., Feigenbaum, L., Singer, D. S. and Singer, A. (2013). Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity 38(6): 1116-1128.
- Vang, K. B., Yang, J., Mahmud, S. A., Burchill, M. A., Vegoe, A. L. and Farrar, M. A. (2008). IL-2, -7, and -15, but not thymic stromal lymphopoeitin, redundantly govern CD4+Foxp3+ regulatory T cell development. J Immunol 181(5): 3285-3290.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Owen, D. L. and Farrar, M. A. (2019). In vitro Differentiation of Thymic Treg Cell Progenitors to Mature Thymic Treg Cells. Bio-protocol 9(16): e3335. DOI: 10.21769/BioProtoc.3335.
Category
Immunology > Immune cell differentiation > T cell
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









