- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visualizing Hypoxia in a Murine Model of Candida albicans Infection Using in vivo Biofluorencence
Published: Vol 9, Iss 15, Aug 5, 2019 DOI: 10.21769/BioProtoc.3326 Views: 5103
Reviewed by: Emily CopeFilipa VazSveta Chakrabarti

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Use of Gas Chromatography to Quantify Short Chain Fatty Acids in the Serum, Colonic Luminal Content and Feces of Mice
Willian Rodrigues Ribeiro [...] Caroline Marcantonio Ferreira
Nov 20, 2018 14584 Views

RNA Extraction from Ears and Draining Lymph Nodes of Mice Infected with Leishmania amazonensis
Emilie Giraud and Evie Melanitou
Jun 5, 2020 5513 Views

TetR Regulated in vivo Repression Technology to Identify Conditional Gene Silencing in Genetically Engineerable Bacteria Using Vibrio cholerae Murine Infections as Model System
Franz G. Zingl [...] Stefan Schild
Oct 5, 2020 3742 Views
Abstract
Candida albicans is a leading human fungal pathogen that uses several metabolic adaptations to escape immune cells and causes systemic disease. Here, we describe a protocol for measuring one of these adaptations, the ability to thrive in hypoxic niches. Hypoxia was generated after successful subdermal infection with C. albicans in a murine infection model. Hypoxia was measured using a fluorescent dye for carbonic anhydrase 9, a host enzyme active under hypoxic conditions. Emitted fluorescence was subsequently quantified using an IVIS system. This protocol was optimized for the use in subdermal infection in mice but has the potential to be adapted to other models of fungal infection.
Keywords: Candida albicansBackground
Fungal colonizers of humans have evolved to sense and adapt to niches available in the host (Grahl and Cramer, 2010). Oxygen is a changing environmental parameter. Levels change in different tissues and during different stages of infection and immune activation (Carreau et al., 2011; Wenger et al., 2015). The ability to sustain growth and to survive in low oxygen environments has been linked to virulence in several fungi (Shepardson et al., 2013; Gresnigt et al., 2016; Pradhan et al., 2018). We have developed a protocol to follow the generation of hypoxia in a mouse infected with C. albicans (Lopes et al., 2018). We chose subdermal infection as a local, non-disseminated model of mycosis with acute onset which allows analysis of hypoxia in a confined space, where C. albicans and host cells interact (Urban et al., 2009; Santus et al., 2018). Thereby, natural variation of oxygen levels in other tissues can be eliminated and secondary effects from distant locations arising in systemic infection can be avoided. During infection, hypoxia is created mainly by neutrophil influx to the site of infection. Neutrophil extravasation and the activation of oxygen-consuming enzymes create environments with low oxygen levels. In turn, C. albicans exploits this environmental shift to avoid immune recognition by changing cell wall composition leading to masking of recognized entities (Lopes et al., 2018).
Materials and Reagents
- Pipette tips (P20, P200, P1000)
- Inoculation loop
- Glass or disposable round bottom sterile tubes for growing 10 ml cultures of yeast (Sarstedt, catalog number: 62.547.004)
- Tube, 50 ml (Sarstedt, catalog number: 62.547.004)
- 1 ml micro-fine U100 insulin syringes (BD, catalog number: 324827)
- Latex gloves
- Candida albicans wt isolate SC5314
- Mice 6- to 8-week-old albino female mice [B6(Cg)-Tyrc-2J/J; Jackson Laboratory-USA]
- PBS (Thermo Scientific, catalog number: 18912014)
- Isoflurane (Orion Pharma, Abbott Laboratories Ltd., Great Britain, CAS: 26675-46-7)
- HypoxiSense 680 Fluorescent Imaging Agent (PerkinElmer, NEV11070r)
- Yeast nitrogen base (Difco, catalog number: 233520)
- Ammonium sulfate (Sigma, catalog number: 31119)
- Yeast Synthetic Drop-out Medium Supplements (Sigma, catalog number: Y1501-20g)
- Glucose (Sigma, catalog number: 47829)
- Synthetic complete dropout medium (SC medium) containing 2% glucose (see Recipes)
- 1x PBS (see Recipes)
Equipment
- Pipettes (P20, P200, P1000)
- 30 °C shaker for growing yeast cultures (speed approximate 180 rpm) (IKA KS 4000i)
- Centrifuge (Eppendorf, model: 5180 R)
- Cell counter Vi-CELL Cell Viability Analyzer Beckman Coulter
- GI-8 gas anesthesia system (Caliper Life Sciences, Inc.)
- Electric clipper to shave mice (Moser Chromini)
- Heating lamp
- Xenogen IVIS Live Animal Imaging System (PerkinElmer)
- Incubator (Binder KB 53)
Software
- Living Image software 4.5 (PerkinElmer)
Procedure
- Fungal growth and mouse preparation (Days 1-3)
- Start C. albicans liquid cultures by inoculation frozen stocks with a loop into 10 ml SC medium broth in a 50 ml tube and incubate for 20 h at 30 °C with shaking at a speed of 180 rpm.
- Take 10 μl of C. albicans culture from the previous day to 10 ml fresh SC medium in a 50 ml tube. Incubate the new mixture for an equivalent time period.
- At Day 3, count the cells in the cell counter and prepare a fresh subculture in 10 ml SC medium with a starting cell number of 5 x 107 cells/ml and let it grow for 3 h. Centrifuge the fungi at 3,000 x g, 10 min at room temperature and re-suspend in 1 ml 1x PBS and count the fungal cell number using a cell counter.
- Adjust the fungal concentration to 5 x 108 cells/ml and keep on ice if not used immediately. Calculation example: To prepare 1 ml of yeast suspension with a concentration of 5 x 108 cells/ml from a stock suspension of 8 x 108 cells/ml, add 625 μl of the stock suspension to 375 μl PBS.
- All mice are injected intraperitoneally with 100 μl HypoxiSense 680 dye (2 nmol/100 μl of HypoxiSense dye per mouse) 24 h before C. albicans subdermal infection.
- Subdermal infection (Day 4)
- Place the mouse in an anesthesia box and regulate the level of isoflurane gas to 2.5% on the GI-8 gas anesthesia system. Remove the mouse from the anesthesia box and gently place the mouse with its nose inside an isoflurane delivering cone.
- With the mouse anesthetized, shave the fur off the dorsal flank using the hair trimmer (Figure 1).
- Pull and hold the mouse skin over the shaved area with two fingers. Insert the syringe into the pulled skin and infect the mouse with 100 μl of the C. albicans inoculum prepared (5 x 107 cells). A small liquid bump under the animal skin will be visible for a few minutes.
- Treat and inject control animals in a similar way using 100 μl 1x PBS.
- Animals should be placed under a heating lamp until they are fully awake to assure their well-being.
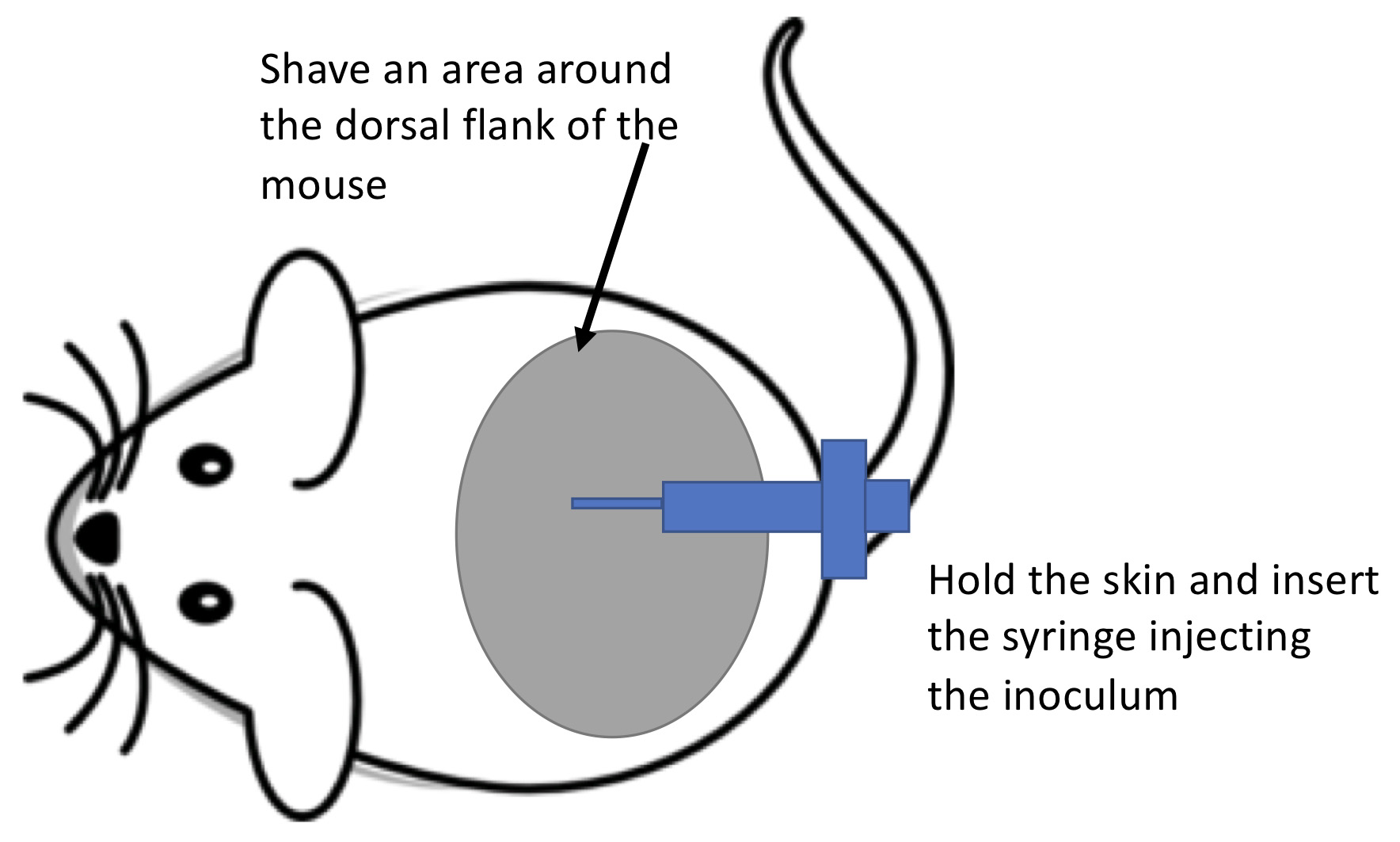
Figure 1. Schematic drawing on how to perform the subdermal infection
- Biofluorescent imaging (Days 4-7)
- Anesthetize the mouse in the anesthesia box using a gas flow of 2.5% isoflurane and transfer them to the camera box of the IVIS imager. Place the mouse facing downwards in the chamber with each animal’s nose inside the cone that delivers the isoflurane.
- Select the IVIS filter setting that fit the emission and excitation wavelength of the dye (675 ex/720 em).
- Using guards to prevent signal bleeding from neighboring animals take a time-zero image of animals in both groups on Day 4 after the initial infection (Figure 2). Expose the animals for 0.5 s exposure capturing in vivo biofluorescence generated by the activity of HypoxiSense dye as a marker for hypoxia. In order to image 5 mice, select D field of view.
- Since the animals might remain under anesthesia for a long period, assure that they are placed under a heating lamp to prevent excessive cooling down of the animal until they are fully awake.
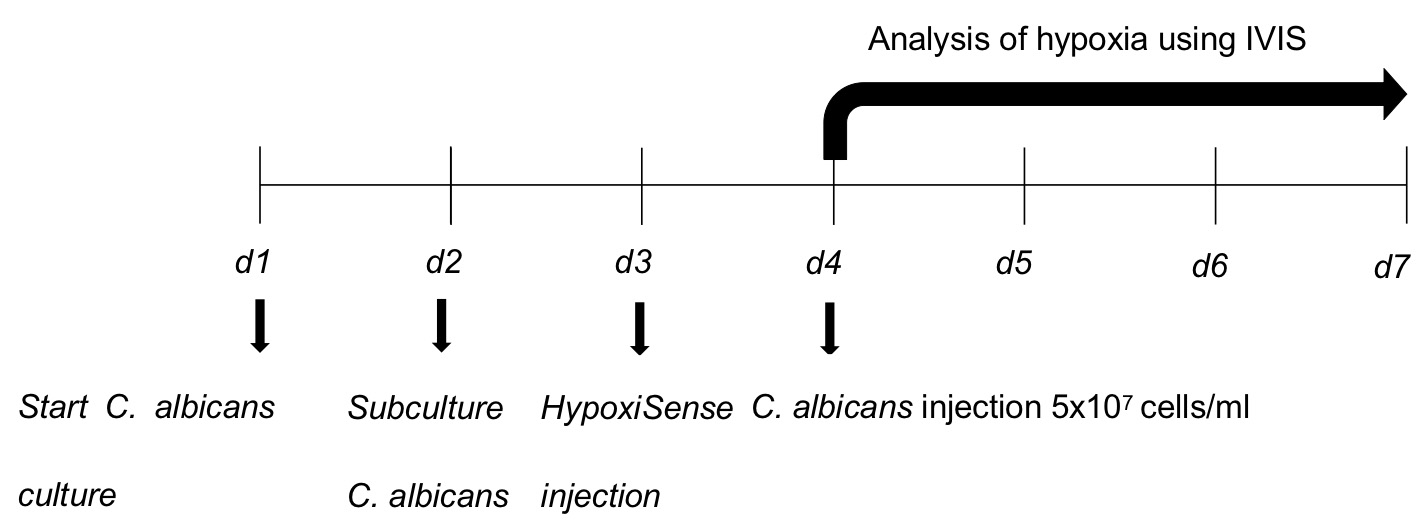
Figure 2. Schematics of the experimental procedure. d: Day.
Data analysis
Image treatment to quantify average epi-fluorescence:
- Draw region of interested (ROI) over the areas of higher epi-fluorescence corresponding to the areas where the abscess is present. Place an additional ROI in a dark region to define a background (Figure 3).
- Perform ROI analyses to measure average epi-fluorescence in each animal.
- Subtract the epi-fluorescence values of the dark background from the values obtained for each animal.
- Use these values to create a graph plot with epi-fluorescence values on the y-scale and the animal group in the x-scale. Statistical significance between the infected and control groups was performed using a two-tailed Mann-Whitney U-test.
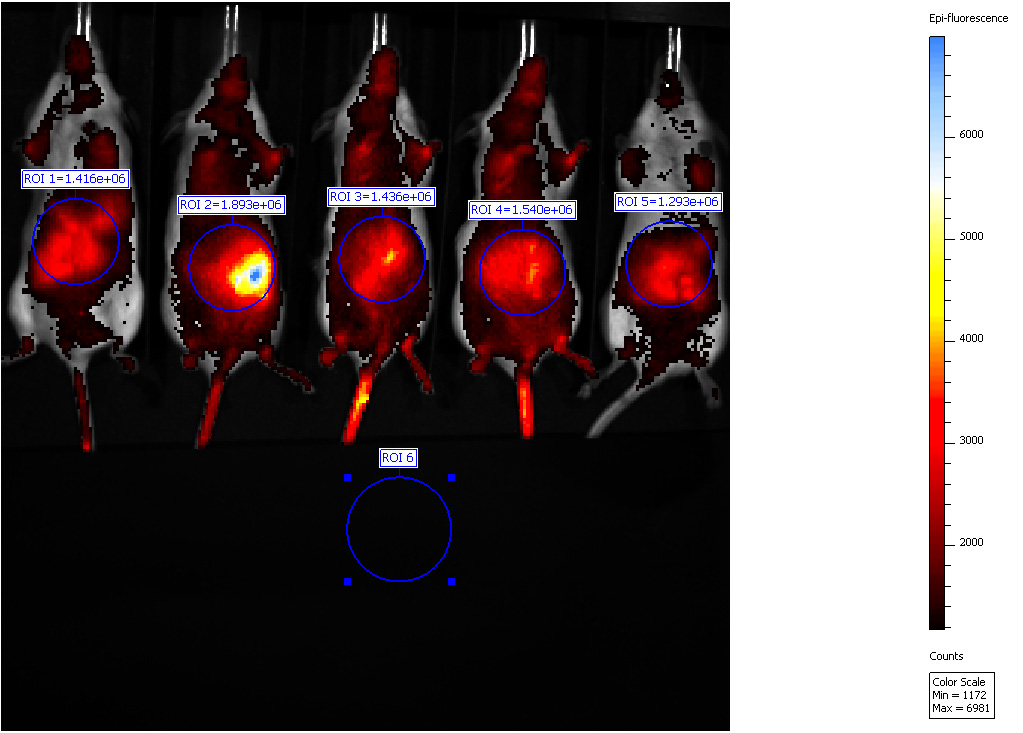
Figure 3. Biofluorescence in vivo imaging. Representative IVIS imaging of mice with hypoxic zones which have developed upon C. albicans infection.
Notes
- This is a non-lethal infection that is self-resolving after 7 days. The fungi do not spread beyond the subdermal tissue and animals appear not be greatly perturbed in their wellbeing (Video 1).Video 1. Mice with a subdermal infection of C. albicans one day after inoculation. The video depicts mice at Day 1 post infection. The mice have received a subdermal injection of C. albicans. The activity and wellbeing of the animals are only affected to a minor extent. (The animal experiments were conducted in accordance with animal ethical guidelines stated in permission A79-14 from the Swedish Board of Agriculture.)
- Animal-to-animal variability and the age of the animal might interfere with the results. Careful selection of the size of the animal groups when planning the experiment is advised. The described protocol used n > 5 animals per group.
- At the end-points animals can be sacrificed and abscess excised to measure infiltration of immune cell or fungal burden.
- Animals were kept at all times in IVC system cages with water ad libitum and monitored daily by the facility care-takers during their stay at the animal infrastructure and by the researchers and the responsible veterinary during the infection period.
Recipes
- SC medium (for 1 L)
Autoclave afterwardYeast nitrogen base 3.4 g Ammonium sulfate 10 g Yeast Synthetic Drop-out Medium Supplements 1.92 g Glucose 2% (w/v) MQ water 1 L - PBS 1x Phosphate-buffered saline (PBS)
Dissolve 1 tablet of PBS in 500 ml MQ water
Autoclave afterward
Acknowledgments
We thank Roland Nordfelth for his assistance with the IVIS equipment. This study was funded by grants to C.F.U. from the Swedish Research Council VR-M 2014-02281 and 2017-01681 and the Kempe Foundation SMK-1453. Additional grants to J.P.L. from J. C. Kempes Minnes Stipendiefond, Arneska Stiftelse, and the Helge ax:son Johnsons Stiftelse are acknowledged. This method is briefly described in Lopes et al. (2018). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Competing interests
All authors declare no competing interests.
Ethics
The animal experiments were conducted in accordance with animal ethical guidelines stated in permission A79-14 from the Swedish Board of Agriculture.
References
- Carreau, A., El Hafny-Rahbi, B., Matejuk, A., Grillon, C. and Kieda, C. (2011). Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15(6): 1239-1253.
- Grahl, N. and Cramer, R. A., Jr. (2010). Regulation of hypoxia adaptation: an overlooked virulence attribute of pathogenic fungi? Med Mycol 48(1): 1-15.
- Gresnigt, M. S., Rekiki, A., Rasid, O., Savers, A., Jouvion, G., Dannaoui, E., Parlato, M., Fitting, C., Brock, M., Cavaillon, J. M., van de Veerdonk, F. L. and Ibrahim-Granet, O. (2016). Reducing hypoxia and inflammation during invasive pulmonary aspergillosis by targeting the Interleukin-1 receptor. Sci Rep 6: 26490.
- Lopes, J. P., Stylianou, M., Backman, E., Holmberg, S., Jass, J., Claesson, R. and Urban, C. F. (2018). Evasion of immune surveillance in low oxygen environments enhances Candida albicans virulence. MBio 9(6): e02120-18.
- Pradhan, A., Avelar, G. M., Bain, J. M., Childers, D. S., Larcombe, D. E., Netea, M. G., Shekhova, E., Munro, C. A., Brown, G. D., Erwig, L. P., Gow, N. A. R. and Brown, A. J. P. (2018). Hypoxia promotes immune evasion by triggering β-Glucan masking on the Candida albicans cell surface via mitochondrial and cAMP-Protein kinase A signaling. MBio 9(6): e01318-18.
- Santus, W., Mingozzi, F., Vai, M., Granucci, F. and Zanoni, I. (2018). Deep dermal injection As a model of Candida albicans skin infection for histological analyses. J Vis Exp(136). Doi: 10.3791/57574.
- Shepardson, K. M., Ngo, L. Y., Aimanianda, V., Latge, J. P., Barker, B. M., Blosser, S. J., Iwakura, Y., Hohl, T. M. and Cramer, R. A. (2013). Hypoxia enhances innate immune activation to Aspergillus fumigatus through cell wall modulation. Microbes Infect 15(4): 259-269.
- Urban, C. F., Ermert, D., Schmid, M., Abu-Abed, U., Goosmann, C., Nacken, W., Brinkmann, V., Jungblut, P. R. and Zychlinsky, A. (2009). Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 5(10): e1000639.
- Wenger, R. H., Kurtcuoglu, V., Scholz, C. C., Marti, H. H. and Hoogewijs, D. (2015). Frequently asked questions in hypoxia research. Hypoxia (Auckl) 3: 35-43.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lopes, J. P. and Urban, C. F. (2019). Visualizing Hypoxia in a Murine Model of Candida albicans Infection Using in vivo Biofluorencence. Bio-protocol 9(15): e3326. DOI: 10.21769/BioProtoc.3326.
Category
Immunology > Animal model > Mouse
Microbiology > Microbe-host interactions > In vivo model > Mammal
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link