- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Solid Phase PCR on 3D Microstructure ArrayChip for Pathogen Detection Application
(*contributed equally to this work) Published: Vol 9, Iss 15, Aug 5, 2019 DOI: 10.21769/BioProtoc.3323 Views: 6359
Reviewed by: Imre GáspárKirsten A. CoprenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid and Multiplex Diagnosis of Malaria Using Chelex-100 Extraction and LAMP-MS Assay
Min Sup Lim [...] Woong Sik Jang
Jul 5, 2025 1838 Views

Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking
Yinwei Gu [...] Haixing Li
Dec 5, 2025 937 Views
Abstract
Advanced free angle photolithography (FAPL) is presented for making 3D supercritical angle fluorescence (SAF) microstructures and transfer them on to polymeric chips using injection molding technique for low-cost microfluidic devices embedded with optical sensing structures. A solid phase polymerase chain reaction (SP-PCR) is used as model technique, which allows rapid and sensitive detection of pathogen DNA on-chip. This article presents the detailed fabrication of SAF structure and SP-PCR application on SAF structure for pathogen detection. This protocol of developing SAF structures using the FAPL process, increases the number of SAF per mm2. FAPL was performed via a motorized stage to control the angle of incidence and to achieve the desired bucket-shapes (dimensions of 50 μm to 150 μm with a slope) required for the 3D optical sensing. Due to the unique properties of SAF structures, it enhances the fluorescent signal by 46 times. Increasing the number of SAF structures and reducing the size resulted in reduction of sample volume required per test along with improvement in the limit of detection (LOD) due to a smaller size. This article also presents the experimental details of SP-PCR using DNA oligos bound to the SAF structures for on-chip pathogen detection and a comparison between different sizes of SAF structures. The direct on-chip SP-PCR paves the path for the application of this technique in point-of-care devices.
Keywords: 3D microstructures arrayBackground
The role of master mold is crucial to generate defect-free polymer chips in an injection molding process (Klank et al., 2002; Hong et al., 2010). This work presents a strategy to maximize on-chip 3D detection sites per mm2 with reduced defects and improved smoothness. Micro milling technique is conventionally used to make a rapid mold for injection molding. Due to the rough surface of milling bits it can cause defects in microstructures during molding. Significant efforts are being made towards improvement of molds by decreasing surface roughness and defects to achieve a better and smoother polymer chip (Ogilvie et al., 2010). Adding the novel FAPL (free angle photolithography) process we report here an improved fabrication of SAF structures and their transfer to the master mold and then to the polymeric chip.
The advantage of using SAF structures can be explained if we consider fluorophore molecule available on the front of a generic surface of microstructure: for a generic structure, most of the emitted light onto the material is refracted outside the structure, and only small amount of light passes through to give a signal (Ruckstuhl and Verdes, 2004; Winterflood et al., 2013). Usually, this refracted fluorescent light is lost or not been gathered when using a flat surface (microscopic polymer slide) for signal capturing. Collection of this remaining part of light by a structure that exhibits supercritical angle of reflection enhances the signal intensity up to 46 times (Hung et al., 2014). SAF structure offers a higher sensitivity and better view for efficient signal collection compared with the flat surface. Fabrication of SAF with photolithography improves the number of structures on a unit area of chip and reduces the volume of sample required. Incorporation of SAF structures on microfluidic chip in combination with SP-PCR provides the capability of multiplexed SP-PCR reactions on-chip for effective low-cost and compact optical detection platform. Our experimental results demonstrated that with SAF array it achieves ultra-sensitive limit of detection (LoD) for fluorophores. This sensitivity is comparable to that obtained by a conventional microarray with data acquisition from a high-end laser scanner. This SAF array platform is advantageous in terms of having a low cost microfluidic platform, whilst retaining high sensitivity and multiplexing capability.
In this paper, we introduce FAPL fabrication process and demonstrate the possibility of achieving lithography-made SAF (L-SAF) structures with lower surface roughness and higher optical efficiency with respect to our previously described micro-milled SAF (M-SAF) (Hung et al., 2015). In particular, we present fabrication procedure (Figure 1) for L-SAF structures of different sizes (50 μm, 100 μm, 150 μm). Multiplexed SP-PCR reaction using DNA probes on L-SAF array for on-chip pathogen detection application (Sun et al., 2011) and our experimental outcomes confirmed that by using these improved SAF arrays, the limit of detection (LOD) could be improved up to 0.05 nM around 6.62 x 103 molecules. The LOD is calculated by data obtained from a high-end fluorescent scanner.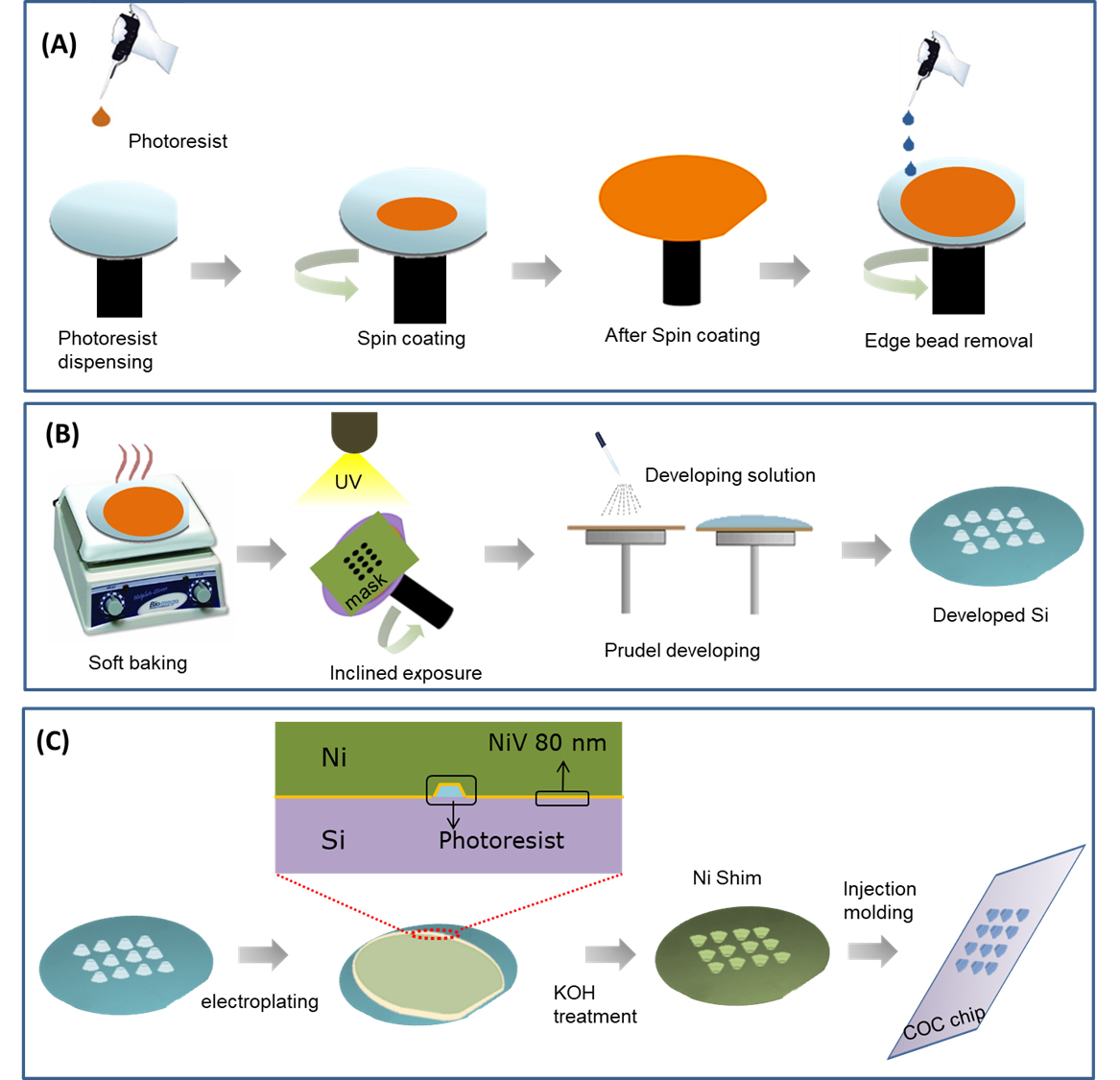
Figure 1. Schematic representation of fabrication steps of L-SAF array using FAPL process. Spin coating of photoresist and an edge bead removal is presented in (A), a free angle UV exposure over the rotatory chuck and development of the exposed Si is presented in (B), the transfer of fabricated 3D L-SAF array to polymeric chip using electroplating of the seed layer of NiV (80 nm) and thick layer (350 μm) of Nickel (Ni) for making a shim for injection molding is presented in (C).
Materials and Reagents
- Glass tip for spotting equipment (PDC 70 Piezo Dispense Capillary, Scienion, Germany)
- Microscope slide (75 mm x 25 mm x 1 mm) (VWR, Denmark, catalog number: 48311-703)
- Microscope slide cover, Gene frame (Thermo Fisher Scientific, Denmark, catalog number: AB0576)
- Negative tone photoresist THB 151N (JSR Micro NV, Belgium), store at cool (4-20 °C) and dark place
- Silicon (Si) wafer (Sigma-Aldrich, Denmark, catalog number: 646687-1EA, CAS Number: 7440-21-3)
- AZ726 MIF developer (MicroChemicals, Germany). Store at cool (4-20 °C) and dark place
- Acetone (Sigma-Aldrich, Denmark)
- TOPAS 5013L COC pellets (Advanced Polymers, Germany), store at room temperature
- DNeasy Blood and Tissue kit (Qiagen, Germany), store at room temperature
- Milli-Q water (Sigma-Aldrich, Denmark), store at 2-8 °C
- Bovine Serum Albumin (Sigma-Aldrich, Denmark), store at 2-8 °C
- Phusion Human Specimen Direct PCR Kit (Thermo Fisher Scientific, Denmark, catalog number: F150BID), store at -20 °C
- Saline Sodium Citrate Buffer, 20x concentrate (Sigma-Aldrich, Denmark, catalog number: SRE0068), store at 2-8 °C
- Triton X-100 (Sigma-Aldrich, Denmark, CAS Number: 9002-93-1), store at room temperature
- Tween-20 (Sigma-Aldrich, Denmark, CAS Number: 9005-64-5), store at room temperature
- Primers (Table 1): purchased from DNA Technology (Aarhus, Denmark)
Note: All primers are diluted in Milli-Q water (Sigma-Aldrich, Denmark) to a final concentration of 100 μM and stored at -20 °C.
Table 1. Primer sequences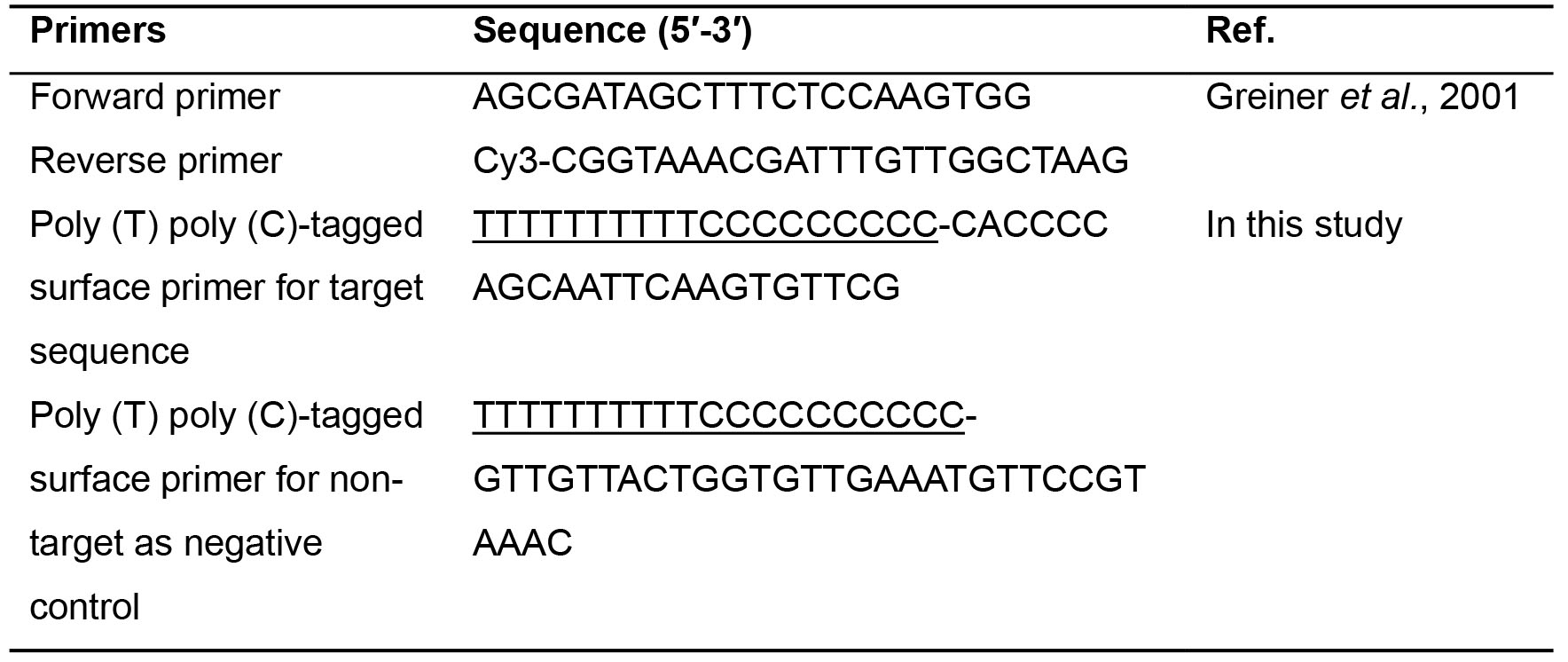
Equipment
- Incubator (Thermo Fisher Scientific, catalog number: 50125590)
- Spin Coater (Karl Suss RCD8, Suss Microtec Germany)
- Hot plate (Thermo Fisher Scientific, catalog number: RC2235Q)
- AXXIS–Co-Sputtering (Kurt J Lesker, England)
- Nickel (Ni) electroplating (Microform.200, Technotrans)
- sciFLEXARRAYER S5 (Scienion, Germany)
- Injection molder (ENGEL Victory 80/45 Tech)
- Custom made rotatory stage for UV exposure (as presented below) (Figure 2)
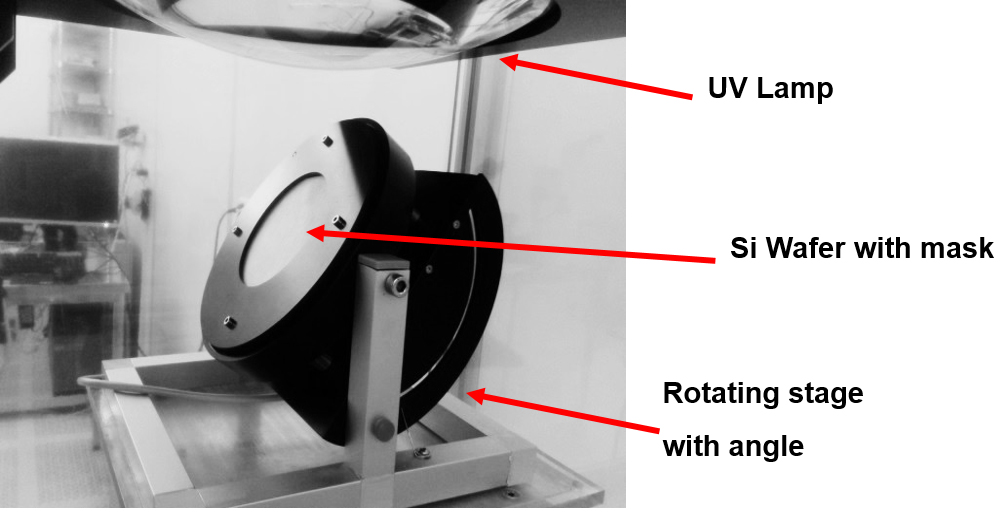
Figure 2. A rotatory stage for free angle photolithography - Si wafers developing system after UV exposure (Gamma 2M Developer, Suss Micro-Tech, Germany)
- Surface profiler (Dektak XTA stylus profiler, Bruker, Denmark)
- BioAnalyzer 4F/4S scanner (LaVision Biotec GmbH, Bielefeld, Germany)
- Nanodrop 1000 (Thermo Scientific, USA)
- ProFlexTM 2x flat PCR System (Thermo Fisher Scientific, USA, catalog number: 4484078)
- Scanning electron microscopy (SEM) (FEI Quanta 200, USA)
Software
- Gwyddion 2.20 analysis software (http://gwyddion.net/download/2.20/)
- ImageJ software (https://imagej.nih.gov/ij/download.html)
Procedure
- Fabrication of 3D microstructure array on Chip
- Take the clean Si wafer and Spin-coat with photoresist polymer (THB 151N at 163.56 x g [1200 rpm]) to obtain a resist layer of about 50 μm thickness.
- To achieve a higher thickness for 100 and 150 μm SAF structures, repeat the spin coating step 2-3 times.
- After spin coating, bake Silicon wafer at 70 °C for 3-5 min.
- The thickness of photoresist coating is chosen to obtain structures with an aspect ratio of about 1, ideal for injection molding.
- Perform edge bead removal to obtain a thickness uniformity under ± 2 μm over the 4-inch wafer area (Figure 1A). The edge bead removal is performed with the robotic arm of the spin coater by dropping continuously acetone solution (to dissolve photoresist) at the edge and continuous spin at 1000 rpm at the same time). Due to the spinning of the wafer, the solution moves outwards so it does not affect the rest of the coating of Silicon wafer.
- Execute the FAPL process with a mask pattern of round holes with diameters ranging from 50 to 200 μm using hard contact mode.
- Load the wafer into the tilting stage, fixed at a given tilt angle and rotated clockwise at 15 rpm and while delivering a dose of 1500 mJ/cm2 throughout the UV exposure.
Note: The photoresist refractive index and the necessity of a 60-degree tilt on the final SAF structures lead to a fixed tilt angle θrot = 52.3 degrees (Figure 1B). - After the UV exposure, automated equipment (Gamma Cluster System 2M, Suss MicroTech) is used to develop the wafers by exposing them to 10 puddles of AZ726 MIF developer for 60 s. Later, rinse the wafers with deionized water before final drying.
- The sputter Nickel Vanadium (Ni/V) = 80 nm or (Chromium (Cr)/ Gold (Au) = 20/60 nm metal Cr/Au seed bilayer together with sputtering 1 μm thick Aluminum (Al) sacrificial layer (Kurt J Lesker) (Figure 1C).
- Perform the Ni electroplating (Microform.200, Technotrans) in three steps by increasing the current in 3 steps. The current goes up linearly from 0 to 0.5 A during the first 15 min, and from 0.5 to 1.5 A for the next 15 min. As soon as the current reaches up to 17 A, keep it constant for 6 h and after that decline it to 0 with a slow rate. The current density should not drop suddenly to minimize the residual stress and achieve a thickness of 350 μm.
- After electroplating, Si is etched out in a KOH bath at 80 °C. Then the remaining Ni shim is cut in required size for installation in injection molding machine (ENGEL Victory 80/45 Tech).
- Use TOPAS 5013L COC pellets for injection molding for polymer chip. The polymers get dry in the hopper of injection molder (airflow at 90 °C) and extra moisture is removed.
- The temperature of injection molding cylinder is increased gradually from 250 °C as polymer exited the hopper to reach 300 °C at the nozzle. Molten COC gets injected into the mold at a speed of 21 cm3/s and a pressure of 600 bars as presented in Video 1.
- The mold temperature may vary (Variotherm process) from 155 °C (at the moment of injection) to 103 °C when the demolding is performed. Also, a packing pressure of injection molding (600 bars) is applied for 6 s injection molded to produce the structures into the microfluidic device. The final chips had the dimensions of a microscope slide (76 mm x 25 mm x 1 mm).Video 1. A schematic video of the Injection molding machine to present the molding process
- Perform the precise spotting on each SAF structure in an array by sciFLEXARRAYER S5 (SCIENION, Germany). Take a glass tip (Type 1) and use it in washing step each time before and after changing the sample for spotting in the software program. This process is automated and controlled by the software developed by SCIENION.
- Solid phase PCR
- DNA preparation
- Streptococcus pneumoniae is from the culture collection of The National Food Institute, Technical University of Denmark (DTU-Food).
- Isolate S. pneumonia genomic DNA using DNeasy Blood and Tissue kit (Qiagen, Germany) as instructed by the supplier.
- Determine DNA concentration with Nanodrop 1000 (Thermo Scientific, USA). Record DNA concentration in ng/μl and quality scores (260/280 and 260/230).
- Store the DNA at -20 °C for further use.
- Report the cut-offs you used for high-quality DNA. The 260/280 ratio of less than 1.7 can be indicative of poor quality DNA. Most studies have cut-offs for 260/280 and 260/230.
- Primers
To make a model to confirm the performance of SP-PCR on FAPL-generated SAF structures for pathogen detection, we selected one pair of a forward and a Cyanine-3 (Cy3)-labeled reverse primer (Table 1) to amplify pneumolysin gene as target sequence for S. pneumoniae detection in the liquid phase (Greiner et al., 2001). A Primer-BLAST from National Center for Biotechnology Information (NCBI) is used to design a nested probe as the surface primer based on pneumolysin gene sequence for SP-PCR. A random sequence from S. pyogenes genome is used as the surface primer for non-target amplification. - Immobilization of surface primers
- Prepare 100 μl solution of surface primer containing 50 μM of poly (T) poly (C)-tagged surface primers containing 5x SSC and 0.004% Triton X-100 at final concentration.
- Spot 1 drop (220 pl) solution of surface primers in front of SAF structure (ф 50 nm) inside the chamber of a polymeric chip (COC) by sciFLEXARRAYER at 20 °C, 65% of humidity.
- Dry microchip at room temperature.
- Treat microchip with UV irradiation at wavelength of 254 nm with the power of 3 mW/cm2 for 10 min (Stratalinker 2400, Stragtagene, CA, USA) to directly immobilize the poly (T) poly (C)-tagged DNA oligonucleotide on the plastic surface without any surface modification (Sun et al., 2012).
- Keep microchip inside a plate containing 0.1x SSC for 5 min, then rinse with Milli-Q water, and dry in an incubator at 37 °C.
- Treat the chip with BSA 2.5 mg/ml for 30 min then rinse with Milli-Q water and dry in an incubator.
- Solid phase-PCR
- Bond and fix the microchip with gene frame (Thermo Fisher Scientific) to create a 25-μl reaction chamber surrounding the solid support primer immobilized SAF array.
- Use a pipettor with a pipette tip to load 25 μl PCR master mix into the gene frame and then seal with a coverslip. The PCR master mix contains:
2 ng of DNA template
1x Phusion® Human Specimen PCR Buffer (Thermo Fisher Scientific)
150 nM of forward and 1,500 nM reverse primers
0.05 U/μl Phusion Human Specimen DNA polymerase (Thermo Fisher Scientific) - Perform the Solid phase-PCR in a ProFlexTM 2x flat PCR System (Thermo Fisher Scientific) with thermal cycling as follows: 94 °C for 3 min, followed by 30 cycles of 94 °C for 20 s, 60 °C for 20 s, 72 °C for 20 s.
- After finishing PCR, wash the chamber with 4x SSC for 5 min, 0.1x SSC containing 1% Tween-20 for 5 min, then rinse with deionized water and dry at room temperature.
- Use a BioAnalyzer 4F/4S scanner with 200-ms shutter time (LaVisionBioTec GmbH, Bielefeld, Germany) to scan chip for data analysis (Figure 3).
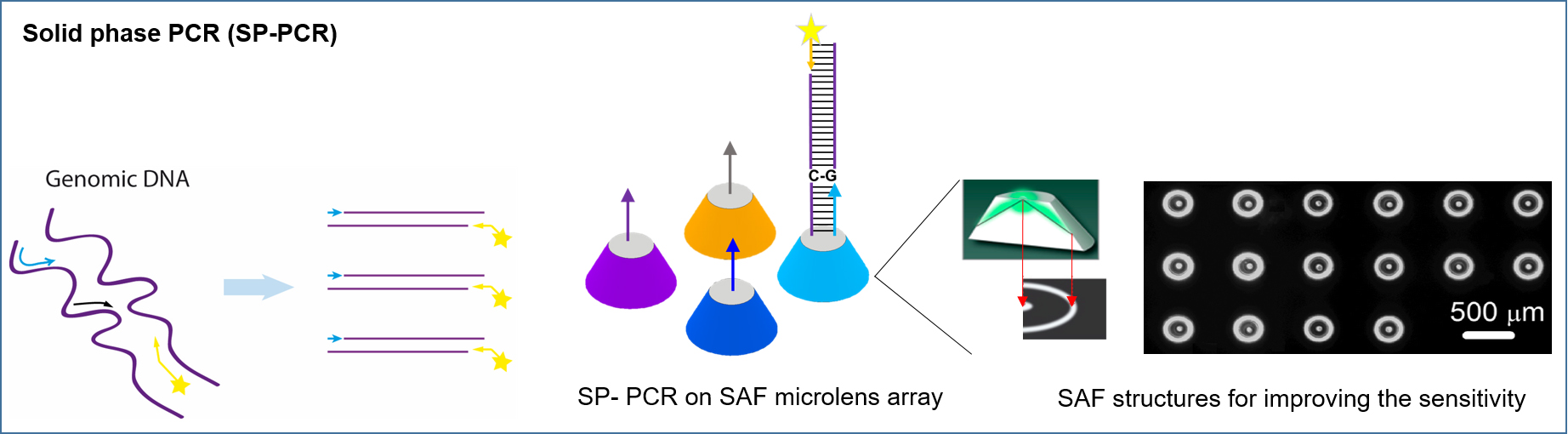
Figure 3. Schematic illustration of SP-PCR technique
- DNA preparation
Data analysis
- Characterization of 3D microstructures
- Perform scanning electron microscopy (SEM) with 3-10 KV beam line under high vacuum condition to characterize the 3D microstructures as presented in Figure 4.
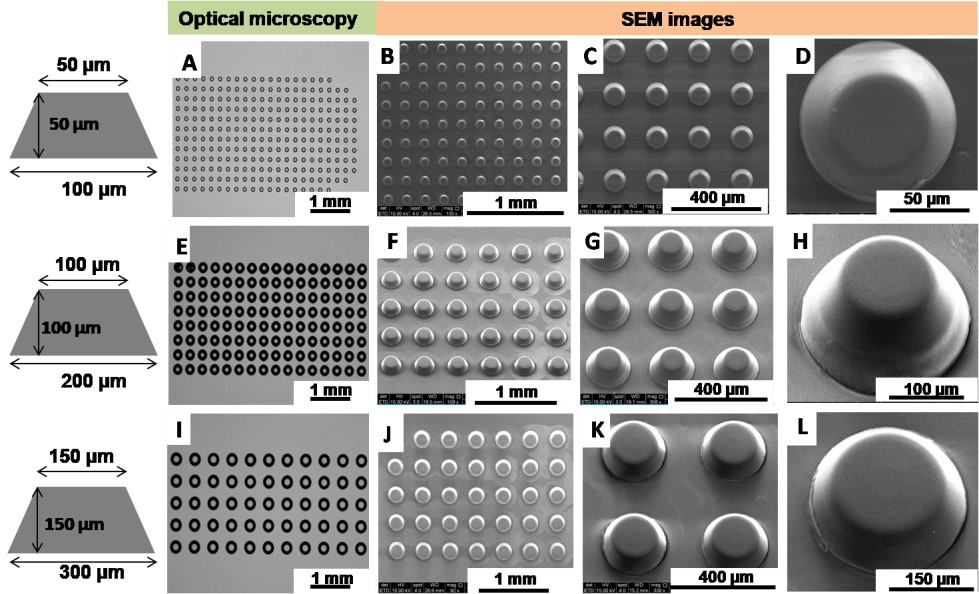
Figure 4. Characterization of fabricated SAF array using the FAPL process using an optical microscope and SEM. (A, E, I) are optical images of SAF structure array. SEM images on the polymeric chip after the transfer of SAF structure by injection molding, for 50 μm size (B, C, D), for 100 μm (F, G, H) and 150 μm (J, K, L). - Use a surface profiler (Dektak XTA stylus profiler, Bruker) and Gwyddion 2.20 analysis software to analyze surface roughness of sidewall surfaces. Figure 5 presents SAF structures with extremely low surface roughness (Ra: 0.5 ± 0.1 μm).
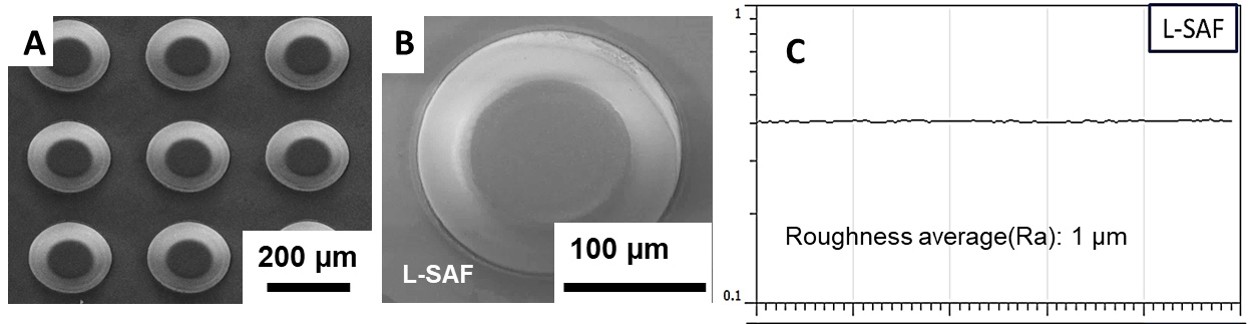
Figure 5. Surface characterization of SAF. SEM images of the SAF structures on polymeric chip after injection molding (A, B) FAPL fabricated SAF (L-SAF) surface profile and average surface roughness of L-SAF (C). - Use the BioAnalyzer 4F/4S scanner with 200-ms shutter time (LaVisionBioTec GmbH, Bielefeld, Germany) to scan microchip after spotting DNA probes or solid phase-PCR. Analyze fluorescence intensity using ImageJ software (Schneider et al., 2012) (Figures 3, 6 and 7). A circle is adjusted to the size of the SAF, and the mean value of grey levels of the pixels inside a fluorescent spot is calculated. A square is drawn surrounding the circle, and the mean signal is taken as background 1. To determine the signal to noise ratio (SNR), the mean of the negative control (NC) fluorescent spots (sample lacking the probe target) is used as background 2 (b2). The SNR in this study is defined as the mean signal intensity of the feature (ave SI) subtracted from the mean background 2 (ave b2) and divided by the variation of the background 2 (stdev b2). Here is the equation: SNR = (AVE(SI)-AVE(b2))/[STDEV(b2)]. The limit of detection was SNR ≥ 3.
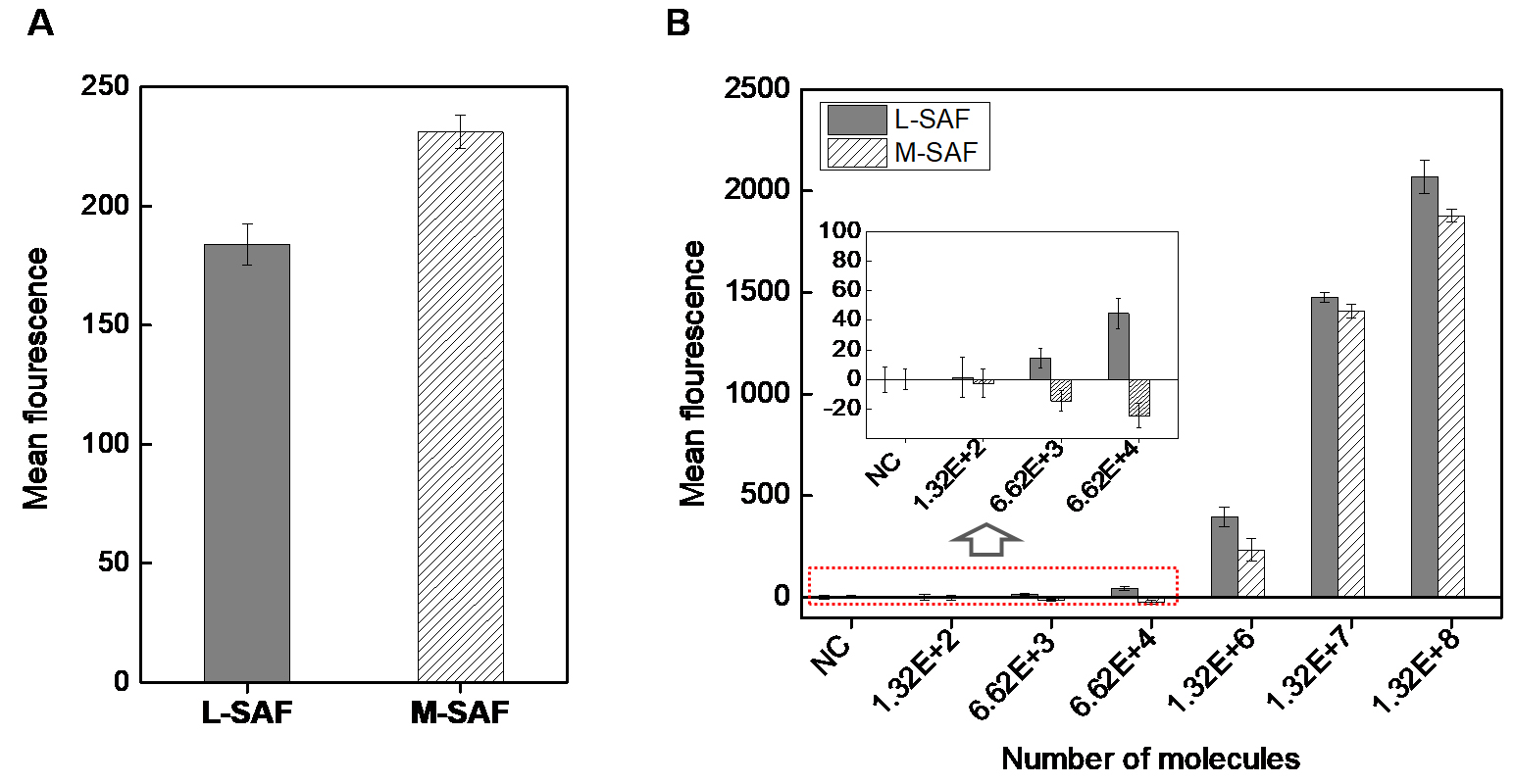
Figure 6. Comparison of fluorescence efficiency, L-SAF vs. M-SAF. A. Comparison of background signal from L-SAF and M-SAF. B. Graph for the mean value of fluorescence signal for L-SAF and M-SAF spotted with different numbers of fluorescent molecules after subtracting background. The spotting volume is 220 pl and exposure time 400 ms for the same size of L-SAF and M-SAF. Error bars of the y-axis were generated as the standard deviation of the mean from at least three replicates.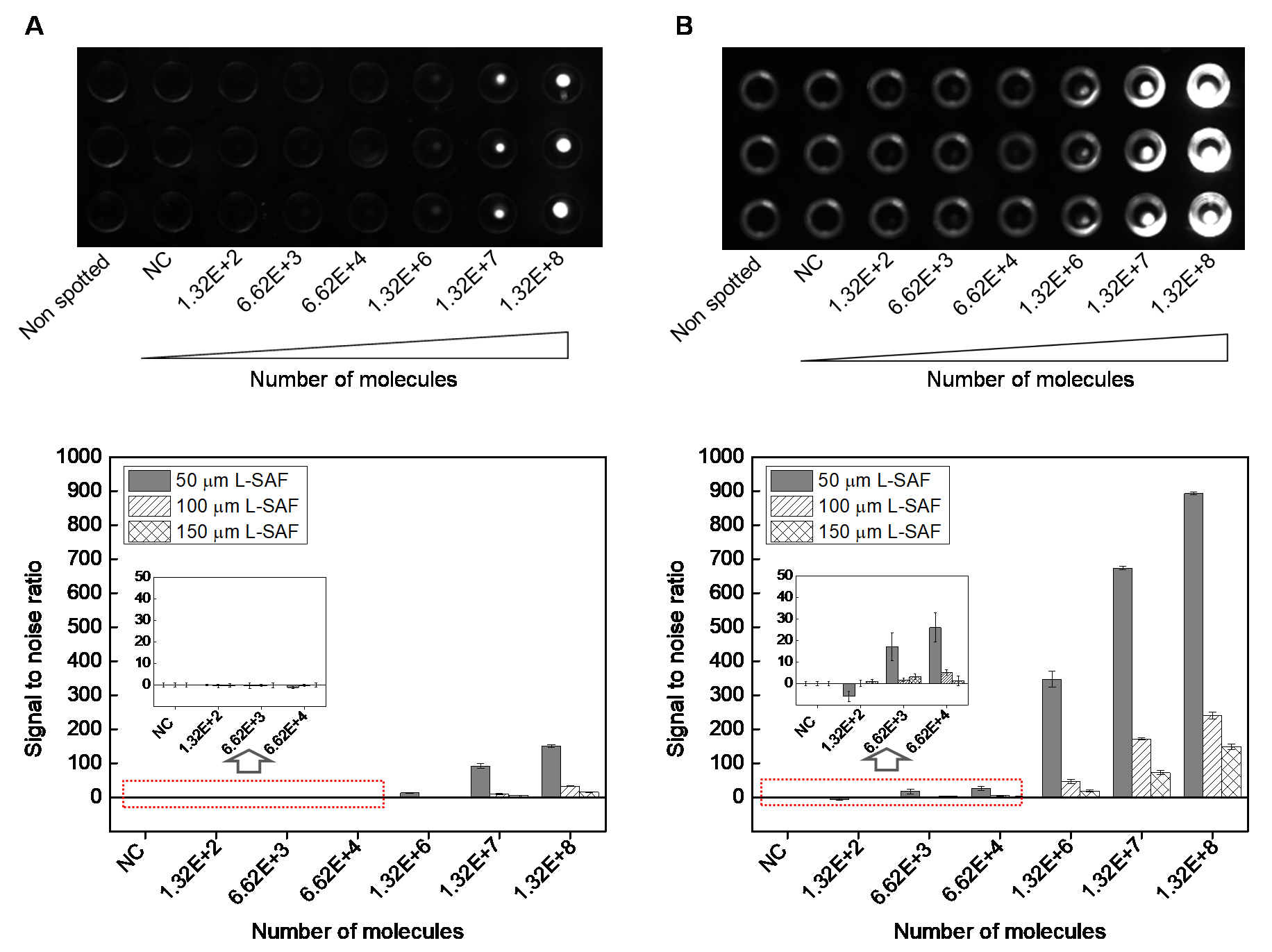
Figure 7. Fluorescent images of L-SAF structures, spotted with 220 pl Cy3-labeled DNA probe ranging from 1 pM (1.32 x 102 molecules/SAF structure) to 1 μM (1.32 x 108 molecules/SAF structure). The images are taken from (A), front side (no use of SAF optical structure) and (B), rear side (use of SAF optical structure). In (B), the light ring corresponded to emission reflected at the side wall of L-SAF structure. Bottom two graphs: signal to noise ratio from 50, 100, and 150 μmL-SAF arrays scanned by using Bio-Analyzer 4F/4S scanner at constant exposure time (400 ms) and with different number of fluorescent molecules per SAF structure. Left graph: scanned from the front side (no use of SAF optical structure), and right graph: scanned from the rear side (use of SAF optical structure). Error bars of the y-axis were generated as the standard deviation of the mean from at least three replicates.
- Perform scanning electron microscopy (SEM) with 3-10 KV beam line under high vacuum condition to characterize the 3D microstructures as presented in Figure 4.
- Evaluation of the SP-PCR
To evaluate the applicability of 3D SAF structure for further biomedical application, SP-PCR (Figure 3) is performed on the front of L-SAF structure embedded in a microchip, where the surface is immobilized by primers as indicated in Figure 8A. DNA target gene (pneumolysin gene) from Streptococcus pneumoniais specifically amplified using SP-PCR on the front of L-SAFs on chip. As shown in Figure 8B, high fluorescent signal and there are no fluorescent signals observed containing non-specific primers.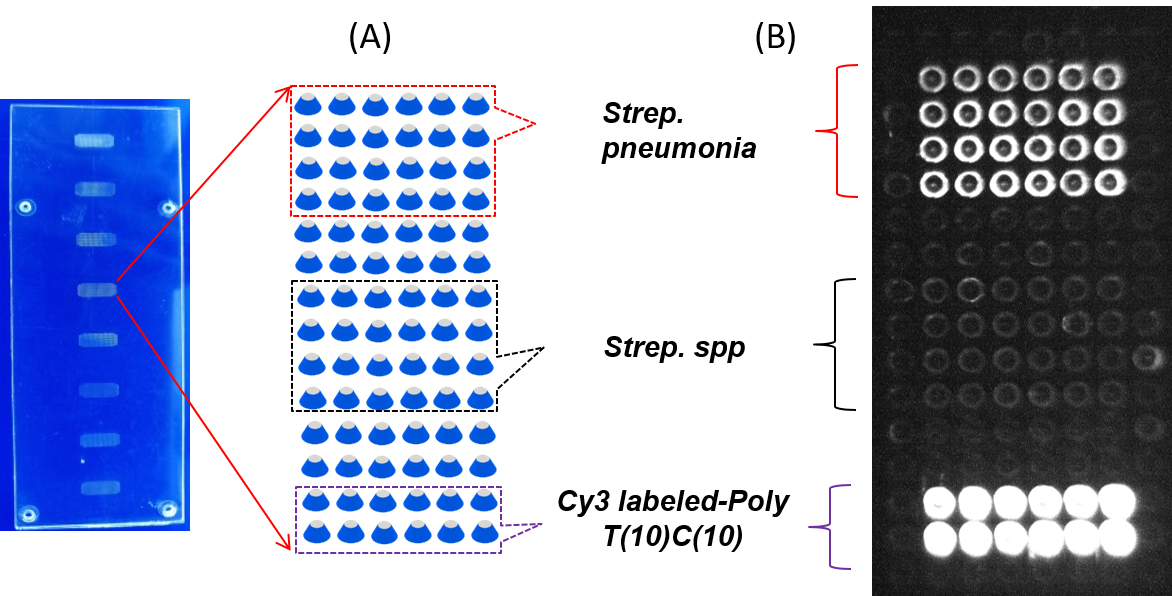
Figure 8. SP-PCR on the L-SAF array. A. Microarray layout. The surface primers for specific amplification of DNA target gene from Streptococcus pneumoniae and Streptococcus spp. and Cy3 labeled-poly T (10) C (10) as a positive DNA probe were immobilized on front of L-SAF for SP-PCR reaction. B. Fluorescent scanning image of the L-SAF array after SPPCR.
Acknowledgments
This work was financially supported by the European Union's Horizon 2020 research and Innovation program, grant agreement No. 687697.
Competing interests
The authors declare no conflict of interest.
References
- Greiner, O., Day, P. J., Bosshard, P. P., Imeri, F., Altwegg, M. and Nadal, D. (2001). Quantitative detection of Streptococcus pneumoniae in nasopharyngeal secretions by real-time PCR. J Clin Microbiol 39(9): 3129-3134.
- Hong, T. F., Ju, W. J., Wu, M. C., Tai, C. H., Tsai, C. H. and Fu, L. M. (2010). Rapid prototyping of PMMA microfluidic chips utilizing a CO2 laser. Microfluid Nanofluidics 9(6): 1125-1133.
- Hung, T. Q., Sun, Y., Poulsen, C. E., Linh-Quyen, T., Chin, W. H., Bang, D. D., Nielsen, L. B., Overby, B., Heller, M. and Wolff, A. (2014). Array of highly sensitive supercritical angle fluorescence micro-optic structures in a disposable lab-on-a-chip for multiplexed detection.18th Int Conf Miniaturized Syst Chem Life Sci MicroTAS: 2244-2246.
- Hung, T. Q., Sun, Y., Poulsen, C. E., Linh-Quyen, T., Chin, W. H., Bang, D. D. and Wolff, A. (2015). Miniaturization of a micro-optics array for highly sensitive and parallel detection on an injection moulded lab-on-a-chip. Lab Chip 15(11): 2445-2451.
- Klank, H., Kutter, J. P. and Geschke, O. (2002). CO(2)-laser micromachining and back-end processing for rapid production of PMMA-based microfluidic systems. Lab Chip 2(4): 242-246.
- Ogilvie, I. R. G., Sieben, V. J., Floquet, C. F. A., Zmijan, R., Mowlem, M. C. and Morgan H. (2010). Reduction of surface roughness for optical quality microfluidic devices in PMMA and COC. J Micromechanics Microengineering 20(6): 065016.
- Ruckstuhl, T. and Verdes, D. (2004). Supercritical angle fluorescence (SAF) microscopy. Opt Express 12(18): 4246-4254.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Sun, Y., Dhumpa, R., Bang, D. D., Hogberg, J., Handberg, K. and Wolff, A. (2011). A lab-on-a-chip device for rapid identification of avian influenza viral RNA by solid-phase PCR. Lab Chip 11(8): 1457-1463.
- Sun, Y., Perch-Nielsen, I., Dufva, M., Sabourin, D., Bang, D. D., Hogberg, J. and Wolff, A. (2012). Direct immobilization of DNA probes on non-modified plastics by UV irradiation and integration in microfluidic devices for rapid bioassay. Anal Bioanal Chem 402(2): 741-748.
- Winterflood, C. M., Ruckstuhl, T. and Seeger, S. (2013). Fast and sensitive interferon-γassay using supercritical angle fluorescence. Biosensors (Basel) 3(1): 108-115.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kant, K. and Ngo, T. A. (2019). Solid Phase PCR on 3D Microstructure ArrayChip for Pathogen Detection Application. Bio-protocol 9(15): e3323. DOI: 10.21769/BioProtoc.3323.
Category
Microbiology > Pathogen detection > PCR
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











