- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Plaque Assay to Determine Invasion and Intercellular Dissemination of Shigella flexneri in TC7 Human Intestinal Epithelial Cells
Published: Vol 9, Iss 13, Jul 5, 2019 DOI: 10.21769/BioProtoc.3293 Views: 6207
Reviewed by: Meenal SinhaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Co-culture of Bacterial and Mammalian Cells to Investigate Effects of Potential Probiotics on Intestinal Barrier Function
Ajitpal Purba [...] Dulantha Ulluwishewa
Jun 20, 2025 2519 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1657 Views

Improved Immunohistochemistry of Mouse Eye Sections Using Davidson's Fixative and Melanin Bleaching
Anne Nathalie Longakit [...] Catherine D. Van Raamsdonk
Nov 20, 2025 1565 Views
Abstract
Shigella flexneri invades the epithelial cells lining the gut lumen and replicates intracellularly. The specialized Type III Secretion System (T3SS) and its effector proteins, encoded on a large virulence plasmid, assist the bacterium to gain access to the cytosol. Thereafter Shigella disseminates to neighboring cells in an epithelial layer without further extracellular steps. Host cell lysis occurs when these bacteria have extensively replicated in the target cell cytosol. Here we describe a simple method to qualitatively as well as quantitatively study the capacity of Shigella to invade and disseminate within an epithelium by assessing the number and size of plaques representing the dead cells in a monolayer of TC7 cells. This classical protocol follows a simple approach of infecting the monolayers of epithelial cell lines with Shigella and visualizing the dead cells as plaques formed against a stained background.
Keywords: Intracellular bacteriaBackground
Shigellosis is a gastrointestinal infection caused by one of four species of Shigella and resulting in about 600,000 deaths annually (Reference 15). Although the infections are reported worldwide, children aged 1-4 years in low- or middle-income countries bear most of the burden of the disease (Kotloff et al., 2018). The bacterium infects the colonic epithelium where it gains access to the cytoplasm and replicates. This intracellular multiplication is followed by dissemination to adjacent cells and modulation of the immune response, eventually leading to cell death and destruction of the colonic mucosa (Ashida et al., 2015). Gene mutations followed by in vitro assays such as the gentamicin protection assay or the plaque formation assay, have led to identification of various virulence factors of Shigella that assist the bacterium in invasion and dissemination. Most of these virulence factors are found on a giant virulence plasmid. Notably, the virulence plasmid encodes the Type 3 Secretion System (T3SS) as well as its effectors and regulators (Puhar and Sansonetti, 2014). The T3SS system is necessary for invasion from the extracellular space. Once in the cytosol, Shigella moves via polarization of actin at one bacterial pole owing to the surface protein IcsA. These actin filaments (so-called “actin comets”) propel the bacteria into adjacent cells, which allows them to disseminate without further extracellular steps (Bernardini et al., 1989). At the molecular level, dissemination is poorly understood, but it is thought to depend on the T3SS, similarly to invasion (Schroeder and Hilbi, 2008).
The plaque assay is a classical functional test to study two important steps in the Shigella infection cycle, invasion events and bacterial dissemination (Oaks et al., 1985). Intracellular replication and dissemination can alternatively be observed by fluorescence microscopy. However, this approach is more laborious and mostly qualitative, not quantitative. Many virulence factors have been identified using the plaque assay as the primary evaluation of the bacterial ability to disseminate to adjacent cells. After initial bacterial entry, the infected cells are covered with an agarose overlay containing gentamicin to eliminate the external bacteria resulting from unproductive spreading into the extracellular medium instead of adjacent cells, thereby ensuring that every plaque corresponds to a single invasion event at the beginning of the experiment. After 48-72 h of incubation, dead cells will have detached and are washed away. The intact cell layer is counterstained with Giemsa, resulting in a clear plaque that is visible against the colored background. The number of plaques gives an estimate of the invasive capacity of the bacteria under study. For example, invasive-deficient Shigella such as the mxiD mutant (Allaoui et al., 1993) or the virulence plasmid cured strain BS176 (Sansonetti et al., 1982) will not form any plaques. On the other hand, the size of plaques reflects the bacteria’s ability to spread within an epithelium. For example, a strain that is fully impaired in dissemination such as virG/icsA mutants (Makino et al., 1986; Bernardini et al., 1989) will show no plaque formation. In contrast, strains that spread more slowly such as the ipgD mutant will lead to the formation of smaller plaques with respect to wild-type bacteria (Puhar et al., 2013; Koestler et al., 2018). This protocol can be used to compare the ability of different Shigella strains, including mutants, to disseminate from the initial site of infection. The protocol, however, is less suitable to assess the invasiveness of bacterial strains, which is readily achieved with the less time-consuming gentamicin protection assay. This protocol is an improvement of the classic plaque assay protocol described by Oaks et al. (1985). Shigella is an enteric pathogen and one of its primary targets are intestinal epithelial cells. While Oaks et al. (1985) and most of the literature thereafter used HeLa cells, an epithelial cell line originating from a cervix cancer sample, TC7 cells are intestinal epithelial cells. The detailed step-by-step nature of this protocol makes it highly reproducible and it can be applied to other Shigella species. Further, it can be modified to extend to other intracellular bacteria such as Salmonella or Listeria, or with other epithelial cell lines.
Materials and Reagents
- Conical flask (Duran, Catalog number: 2121628)
- Microfuge tubes (Eppendorf, Safe-Lock tubes, catalog numbers: 0030120086, 0030120094)
- Centrifuge tubes (Sarstedt, catalog numbers: 62.554.502, 62.547.254)
- Culture tube (TPP, catalog number: 91016)
- Pipette tips (VWR, catalog numbers: 89041-404, 89041-412, 89041-400)
- Serological pipettes (VWR, catalog numbers: 612-3702, 612-3700, 612-3698)
- 6-well tissue culture test plates (TPP, catalog number: 92406)
- Cell counter slides (Countess Cell Counting Chamber Slides, Thermo Scientific, catalog number: C10228)
- TC7 cells (Chantret et al., 1994)
- Shigella flexneri M90T (Sansonetti et al., 1982), can also be purchased from ATCC and available on request from the authors
- Dulbecco's Modified Eagle Medium (DMEM) (Gibco, catalog number: 21885-025)
- Penicillin-Streptomycin solution (10,000 U/ml) (Gibco, catalog number: 15140122)
- Minimum Essential Medium Non-Essential Amino Acids solution (100x) (Gibco, catalog number: 11140-035)
- Dulbecco’s Phosphate Buffered Saline (PBS) solution (Gibco, catalog number: 14190-144)
- Fetal Bovine Serum (Gibco, catalog number: 10500056)
- HEPES 1 M solution (Gibco, catalog number: 15630-080)
- Trypsin-EDTA (0.05%), phenol red (Gibco, catalog number: 25300-054)
- Trypan Blue Stain (0.4%) (Thermo Scientific, catalog number: T10282)
- Gentamicin sulfate (Sigma, catalog number: G1264)
- Agarose (VWR Life Science, catalog number: 35-1020)
- Tryptic Soy Broth (TSB) ready to use powder (Merck, catalog number: 105459)
- Tryptic Soy Agar (TSA) ready to use powder (Merck, catalog number: 105458)
- Congo red (Sigma, catalog number: C6277)
- Ethanol (VWR chemicals, catalog number: 20821.558)
- Giemsa's azur eosin methylene blue solution (Merck, catalog number: 109204)
- Growth medium (see Recipes)
- Infection buffer (see Recipes)
- Agarose overlay (see Recipes)
- 5% (v/v) Giemsa solution (see Recipes)
- Congo red solution (see Recipes)
Equipment
- Biosafety cabinet (Thermo Scientific, HERAsafe KS 18)
- Spectrophotometer (Amersham, Ultrospec 2100 pro)
- Pipetteman (VWR, Accurpette)
- CO2 Incubator (Thermo Scientific, Heracell VIOS 160i)
- Benchtop centrifuge (VWR, Micro Star 17R)
- Centrifuge (Eppendorf, 5810R with Rotor A-4-81 for plates)
- Water bath (Grant, JBA 12)
- Pipettes (Eppendorf, Research plus)
- Inverted microscope (Motic, AE2000 Binocular)
- Cell counter (Thermo Scientific, Countess II FL)
- Imaging system (Bio-Rad, ChemiDoc XRS)
- Microwave oven (Whirlpool, MD101)
- Disposable scalpel (Swann-Morton, 0503)
- Vacuum pump (VWR, Mini diaphragm vacuum pump VP 86)
- Orbital shaker (Edmund Bühler, Swip SM25)
Software
- Prism 7 (GraphPad, https://www.graphpad.com)
- ImageJ (https://imagej.nih.gov/ij/index.html)
Procedure
Note: The experiment should be carried out in a biosafety level 2 Lab.
- Preparation of bacteria
Day 1- Prepare or melt tryptic soy agar (TSA), as per manufacturer’s instructions, and let it cool down to about 45°C (e.g., in a water bath already set at 45 °C). Add sterile Congo red solution (see Recipes) to a final concentration of 0.01%, e.g., 5 ml of Congo red solution to 500 ml of TSA. Pour the plates (about 25 ml of TSA in each 10 cm diameter plate) next to an open flame or in a sterile cabinet and let them dry. Store the plates at 4°C and use them within 3 weeks.
- In the afternoon, streak out Shigella flexneri from the glycerol stock (see Notes) on a TSA plate supplemented with Congo red. Grow overnight (approximately 18 h) at 37 °C.
- Inoculate 5 ml of TSB with a single red colony (Figure 1) of Shigella flexneri and incubate overnight at 37 °C with 180 rpm agitation.
- Dilute the overnight culture 200 times in fresh TSB in a conical flask. For instance, in a 125 ml flask, take 25 ml of TSB and add 125 µl of the overnight culture. Incubate at 37 °C with 180 rpm agitation and grow till mid log-phase (OD600nm = 0.3-0.4). The doubling time of Shigella during the exponential phase is about 40 min. It typically takes about 3 h to reach the mid log-phase.
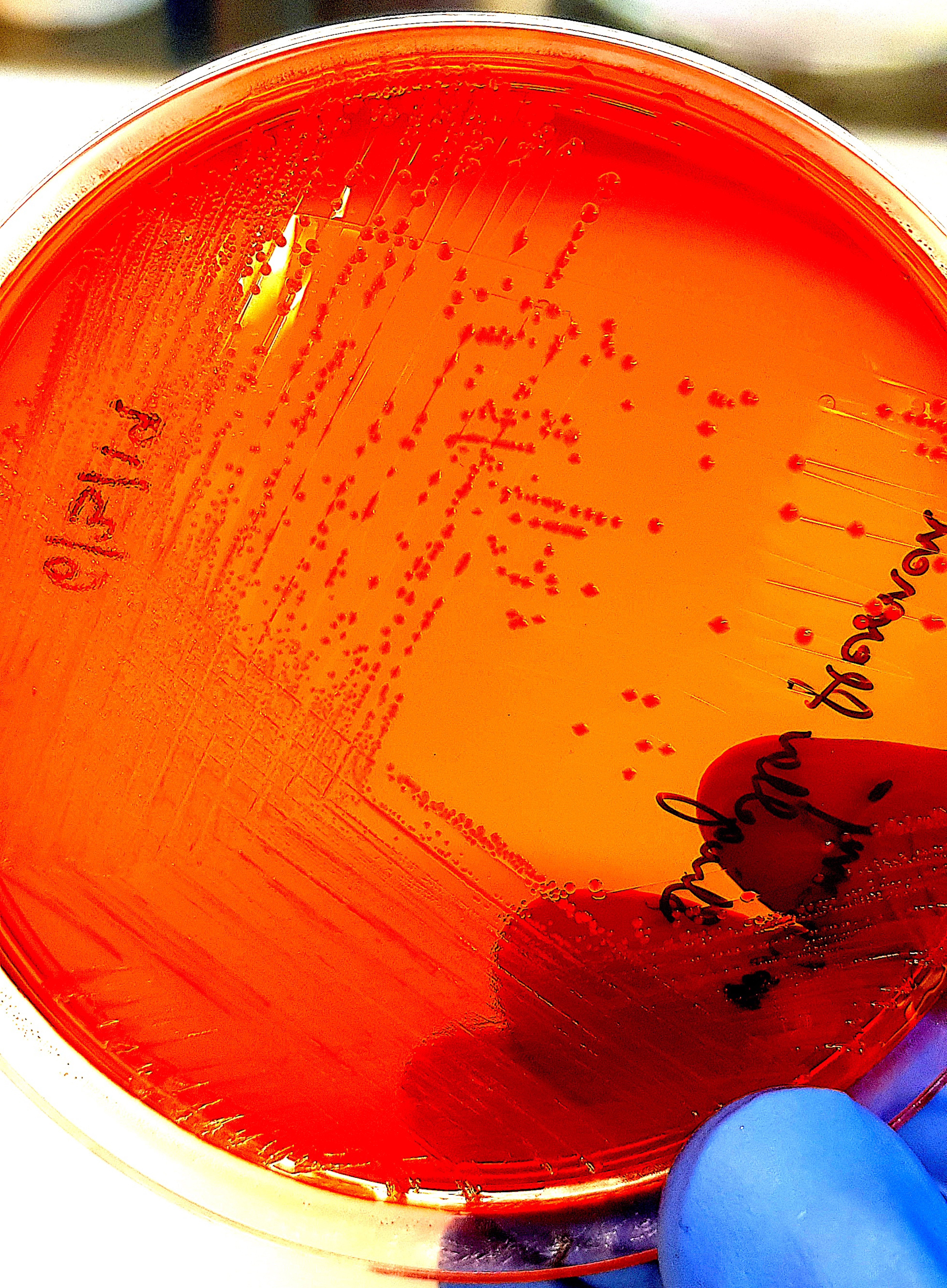
Figure 1. Characteristic red colonies of Shigella flexneri on TSA supplemented with 0.01% Congo red
- Preparation of TC7 cells
Day 2- Remove the growth medium (see Recipes) from a cell culture flask with a confluent monolayer of TC7 cells.
- Wash the cells with sterile PBS three times.
- Add 5 ml of pre-warmed Trypsin solution and incubate the flask at 37 °C for about 4 min.
- Add 5 ml of growth medium (see Recipes) to inhibit the Trypsin and collect the trypsinized cells in a 50 ml centrifuge tube.
- Centrifuge at 900 x g and discard the supernatant. Re-suspend the pellet in 5-10 ml of growth medium devoid of antibiotics (see Recipes) by gentle pipetting.
- In a sterile 1.5 ml microcentrifuge tube, mix 10 µl of cell suspension with 10 µl of Trypan blue solution. Load 10 ml of this mixture on to the Countess II counting slide and using a Countess II cell counter, determine the number of cells.
- Seed the required number of wells in a 6-well plate with 1 million cells (1 x 106) per well, adding at least 2 ml of growth medium devoid of antibiotics (see Recipes) in each well. This number of cells is enough to form a fully confluent monolayer after one day of growth (Figure 2).
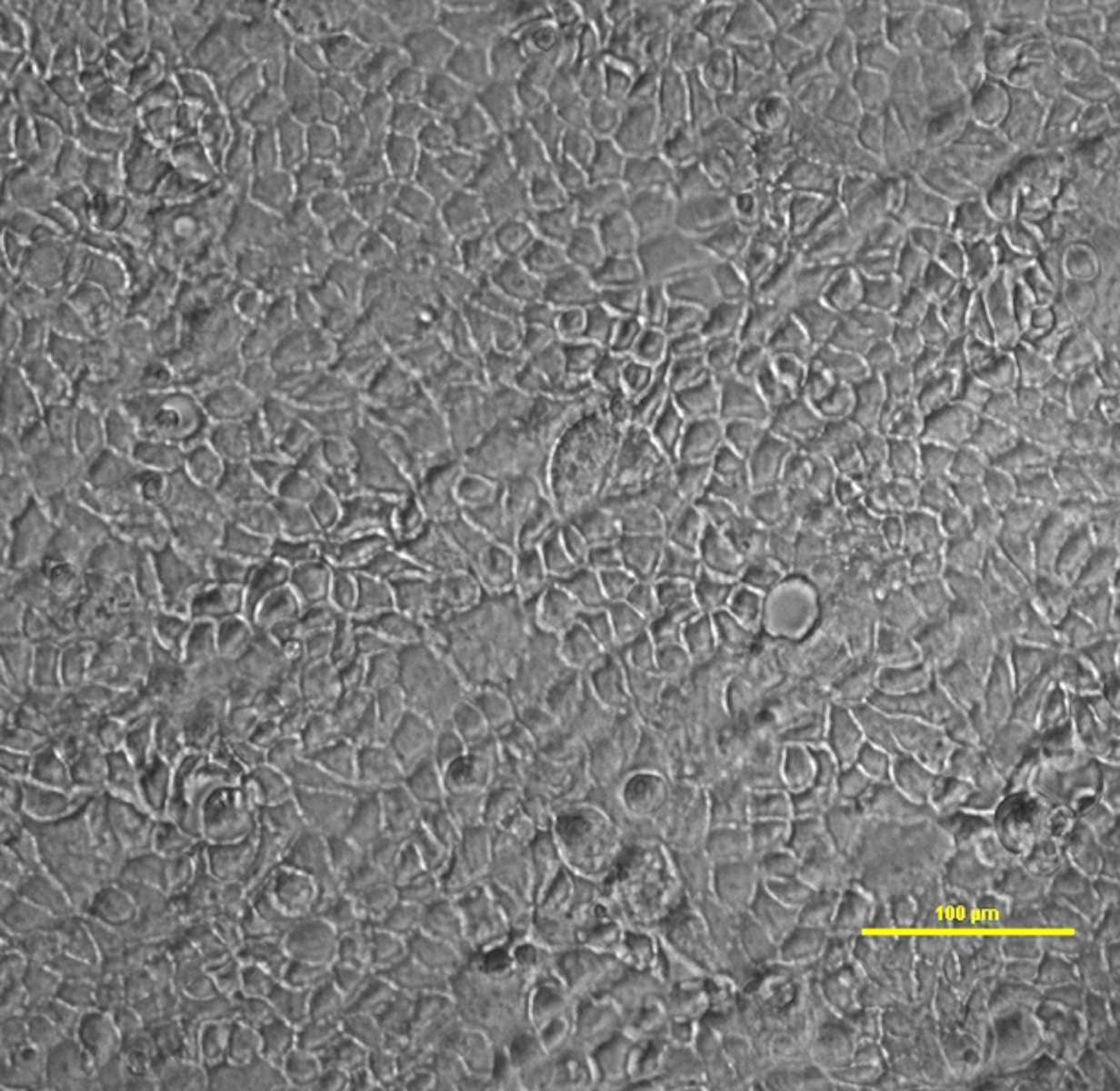
Figure 2. Bright-field image of a monolayer of TC7 cells
- Infection
Day 3- On Day 3, when the bacteria reach OD600nm = 0.3-0.4, pipette 1-1.5 ml culture into a microcentrifuge tube.
- Spin the tube at 3,000 x g for 5 min. Aspirate the supernatant. Add an equal volume of PBS at room temperature and resuspend the pellet by pipetting up and down or by gentle vortexing.
- Repeat Step C2 at least twice and finally resuspend the pellet in infection buffer at room temperature. Record the OD600nm of the bacterial suspension.
The recommended multiplicity of infection (MOI) for this experiment is very low, 1 bacterium for every 150 cells. Since there are 1 x 106 cells in each well, the number of bacteria required per well is about 6,700.
The number of Shigella cells at OD600nm = 1 is 0.5 x 109/ml; hence at OD = z, the number of bacterial cells = z x 0.5 x 109. Calculate the volume of bacterial suspension required for infecting each well using the following formula:
Volume of bacterial suspension required (in ml) = 6,700/z x 0.5 x 109
Additionally, make 10-fold serial dilutions of the bacterial suspension and spread 100 µl of each on TSA plates. Incubate the plates at 37 °C and count the colonies obtained the next day. This colony count of the inoculum will verify the number of bacteria used. - Aspirate the medium from the TC7 cells plate and add 2 ml of PBS at room temperature to each well.
- Swirl the plate gently and carefully aspirate all the liquid. Wash the cells twice in a similar fashion and add 2 ml of infection buffer (see Recipes) at room temperature to each well.
- Add the desired volume of bacterial suspension (calculated in Step C3) to each well of the 6-well plate. Mix well by gentle pipetting to ensure that the bacterial suspension is mixed uniformly. Centrifuge the plate (using the plate adaptors in the centrifuge rotor) at 180 x g for 10 min at room temperature.
- Incubate the plate at 37 °C, 10% CO2, for 2 h.
- After incubation, wash the wells with prewarmed PBS three times as in Steps C4 and C5.
- Completely aspirate the liquid from the wells and cover cells with 2 ml of the cooled agarose overlay (see Recipes).
- Put the plates back at 37 °C, 10% CO2. Examine the plates daily for appearance of plaques. Two days incubation can be enough for optimal plaque size (1-2 mm diameter). However, it is dependent on the bacterial strain and frequent observation is therefore required.
- Visualization and enumeration of plaques
Day 5 or 6- After 48-72 h of incubation, very carefully peel off the agarose overlay using a scalpel to detach the overlay from the walls and by turning around the plate.
- Fix the cells by incubation in 95% ethanol for 5 min.
- Remove the ethanol and stain the wells with 5% Giemsa solution (see Recipes) for 10 min.
- Aspirate the stain and wash the wells twice with distilled water. Let the plate air-dry.
- When the plate is dry, take a picture of the plate using an imaging system (Figure 3).
- Count the number of plaques formed in each well with a marker pen.
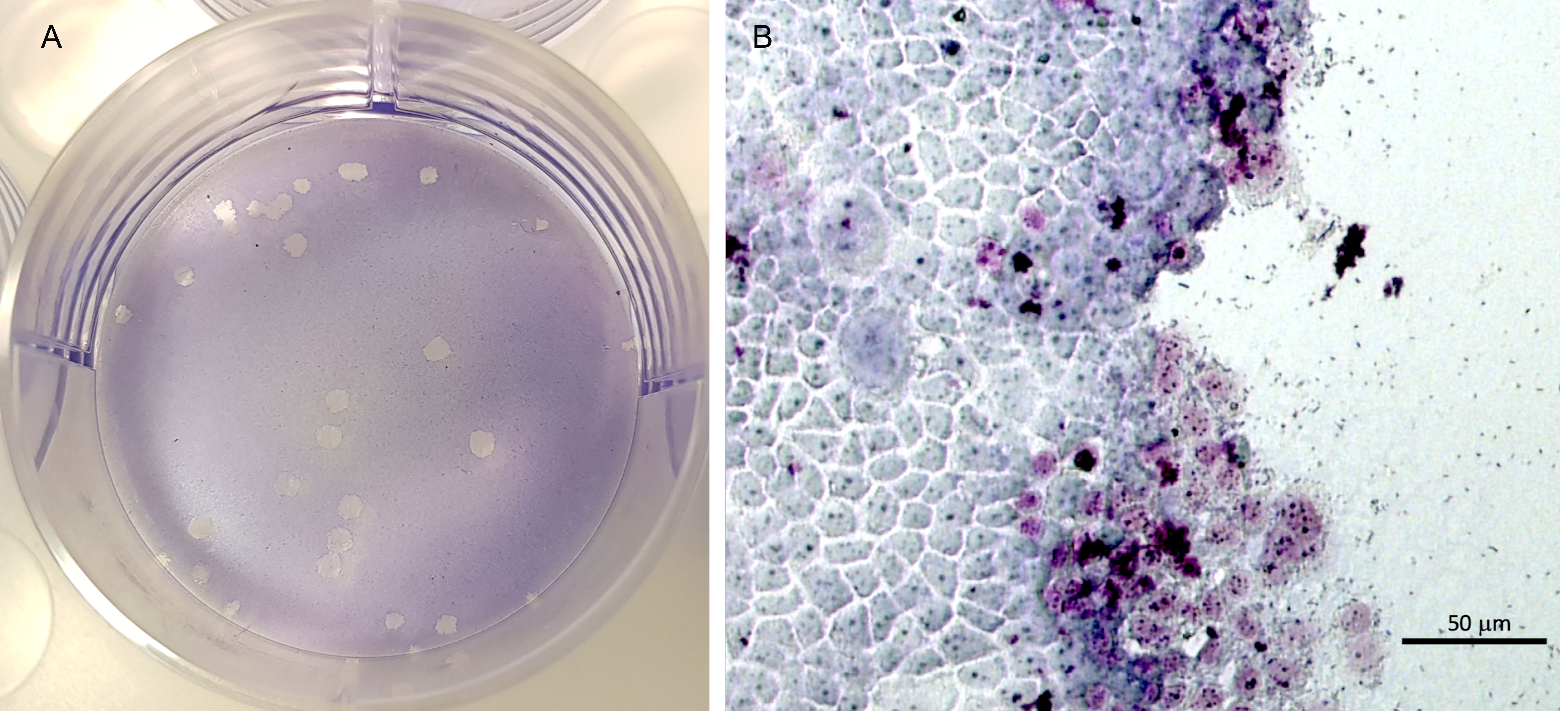
Figure 3. Visual assessment of the plaques formed on the cellular monolayer. A. Clear plaques representing dead cells in a well of 6-well plate with TC7 monolayer stained with Giemsa. B. Bright-field micrograph of a plaque edge. The clear zone is the plaque and the interface of plaque and the TC7 monolayer shows characteristic damage to the cells.
Data analysis
- Using the ImageJ software determine the diameter of each plaque in the image.
- Using the Prism software plot the number or the average diameter of plaques against the condition (e.g., the strain) or the MOI used in particular well (Figure 4). For every condition tested, prepare at least duplicate samples in every experiment to serve as the technical replicates. For biological replicates, repeat the experiment on another day with the same number of bacteria and cells at least 3 times.
- To compare only two sample values at a fixed time point, use Student’s t-test. However, to assess the statistical significance of multiple samples across multiple time points, use two-way ANOVA with Tukey’s post-hoc test.
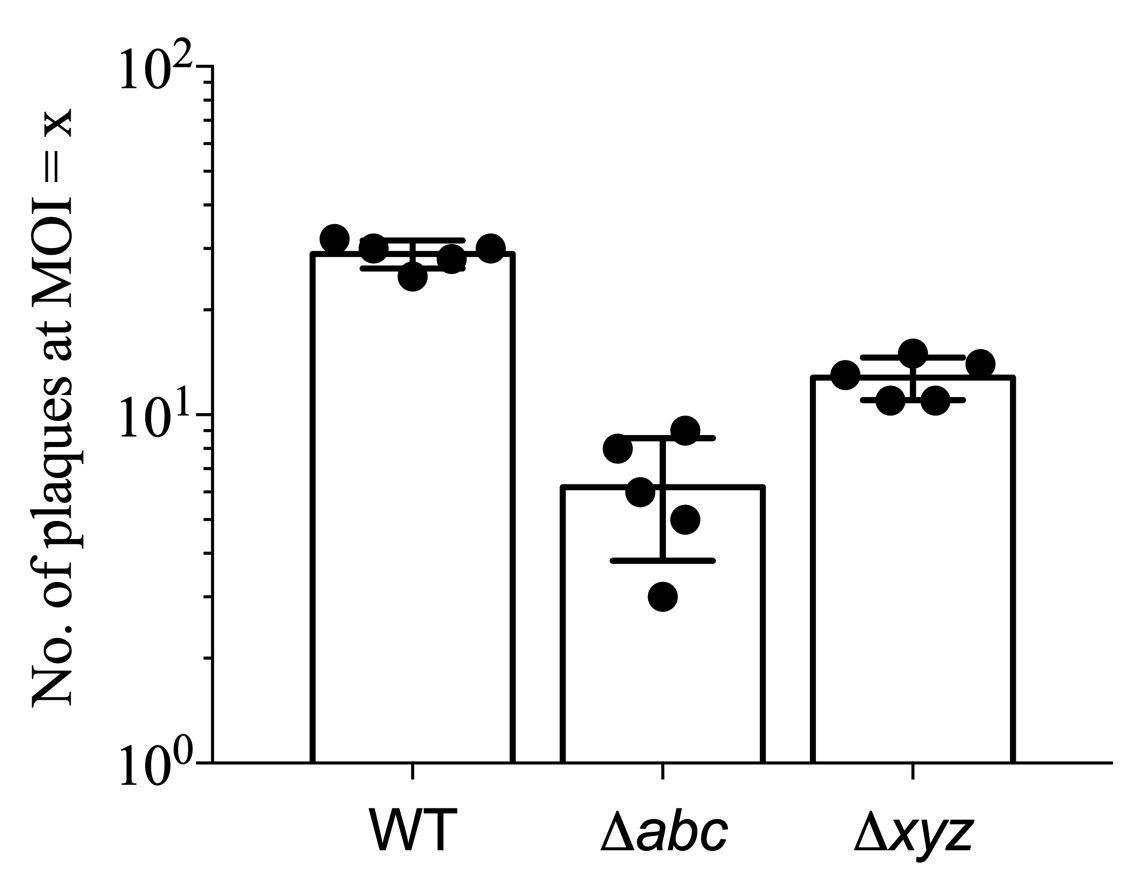
Figure 4. An example plot of the number of plaques at a particular MOI for different bacterial strains. Each dot represents an independent value obtained from a replicate.
Notes
- To prepare glycerol stocks of Shigella flexneri, dilute the overnight culture 200 times in fresh TSB and incubate at 37 °C with 200 rpm shaking. Monitor the OD600 of the culture periodically. At OD600 = 0.6, withdraw 1 ml of culture in a 2 ml cryovial and add sterile glycerol to a concentration of 15-50% (v/v). Mix gently and store the tube at -80 °C. The frozen stocks are stable for years at -80 °C, however, multiple freeze-thaw cycles may reduce the shelf life.
- Carefully select only the red colonies on Congo red agar for the experiment. The off-white colonies have lost the virulence plasmid and are invasion-deficient. If plates are not used immediately after overnight culture, but kept at 4 °C (which is possible for up to 3 weeks), white colonies need to be marked because they will turn red due to unspecific binding of Congo Red. Red staining of invasive colonies only develops during growth above 35 °C.
- Congo red is able to induce secretion via the Type III Secretion System (T3SS) in Shigella. The dye binds to the bacterial colonies that have an active T3SS making them appear red in color. It is a simple and quick screening method to differentiate virulent Shigella colonies from the avirulent ones (due to curing of the virulence plasmid). Unfortunately, the mechanism of action of Congo red is not understood.
- The TC7 cells are cultured and maintained in presence of Penicillin and Streptomycin (100 U/ml). However, infections are carried out in infection buffer that does not contain any antibiotics (which may inhibit the bacterial growth). Ideally, cells are seeded in medium devoid of antibiotics if used the next day or at least extensively washed at the beginning of the experiment.
- The multiplicity of infection (MOI) for this experiment is to be kept very low, because neither the plaque number nor the plaque size can be determined properly when plaques are too close to each other. The MOI can be empirically determined by carrying out experiments with various dilutions of the bacterial suspension. We recommend one bacterial cell per 150 or 300 TC7 cells.
- The 6-well plates are centrifuged after addition of bacteria to aid in binding as Shigella lacks adhesion factors.
- The agarose overlay should not be hot while pouring. It may kill the cells.
- Even during longer incubations, gentamicin won’t be able to penetrate the cells and kill the intracellular bacteria. The intracellular bacteria remain ‘protected’ all the time.
- Keep all the buffers and reagents that are required at the initiation of invasion and thereafter, warmed up to 37 °C before using. The secretion of T3SS effectors mediating invasion is temperature dependent. It is active above 35 °C, but inactive at room temperature. In order to synchronize the invasion, it is important to keep the bacteria in buffers at room temperature before invasion.
- All the reagents can be prepared on Day 1, except for the agarose overlay and Giemsa solution, which must be prepared freshly.
- When using the pipette controller, gently dispense the solutions with gravity (“G” setting in Accurpipette) or at the lowest flow speed into the 6-well plate to avoid detaching the cell monolayer.
- Be careful while swirling the plate, washing the cells and during centrifugation, because the confluent cell monolayer detaches easily.
- The average number and diameter of the plaque cannot be substituted with the measure of total plaque area. The number of plaques actually reflects the number of invasion sites. The diameter of the plaque reflects the capacity of the bacterium to disseminate. Therefore, total plaque area measurement won’t allow distinguishing between a bacterial strain that is poorly invasive but good at disseminating from a strain that is proficient in invasion but is poor at disseminating.
- For further reading and details on basic methods that have not been described in this manuscript (such as bacterial inoculation, pouring the plates, etc.) the reader is referred to the following books: Microbiology: A Laboratory Manual (Cappuccino and Welsh, 2017) and Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications (Freshney, 2010).
Recipes
- Congo red solution
1% (w/v) Congo red in water
Weigh 1 g of Congo red dye and dissolve it in 70 ml of water. Bring the volume of the solution to 100 ml and filter sterilize the solution, using 0.2 µm membrane filters. The Congo red solution is stable for a long time at room temperature - Growth medium
DMEM
1x amino acid solution
100 U/ml Pen-Strep solution
10% heat inactivated FBS
To 500 ml DMEM, add 5 ml of Pen-Strep solution, 5 ml of amino acid solution and 50 ml of FBS (inactivated by incubation at 55 °C for 30 min)
Note: The growth medium must be stored at 4 °C and can be used for 3-4 weeks. - Infection buffer
DMEM (serum free)
20 mM HEPES, pH 7.4
To 50 ml of serum free DMEM add 1 ml of 1 M sterile HEPES pH 7.4
Note: Infection buffer is stable at 4 °C for months. - Agarose overlay
0.5% (w/v) agarose in infection buffer
5% FBS
50 µg/ml gentamicin- Add 0.1 g of agarose to 20 ml of infection buffer
- Melt the agarose in a microwave oven
- When it cools down to about 50 °C, add 1 ml of heat-inactivated FBS and 20 µl of 50 mg/ml gentamicin solution
- Allow the agarose to cool down to about 45 °C in a water bath, so that it is still liquid but not too hot
- 5% (v/v) Giemsa solution
Prepare a 20 ml working Giemsa solution by using 1 ml of stock Giemsa solution to 19 ml of PBS
Keep it in a dark bottle/tube
Note: Do not store it and always use it as a freshly prepared solution.
Acknowledgments
We thank members of the Puhar lab for critical reading. A.S. is the recipient of a stipend from The MIMS Excellence by Choice Postdoctoral Programme under the patronage of Emmanuelle Charpentier. The programme is financed by the Kempe Foundations and the Knut and Alice Wallenberg Foundation. A.P. acknowledges generous funding from MIMS, UCMR, Umeå University and the Wallenberg Academy Fellow programme. This protocol was used in Puhar et al. (2013).
Competing interests
The authors declare no conflict of interest.
References
- Allaoui, A., Sansonetti, P. J. and Parsot, C. (1993). MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol Microbiol 7(1): 59-68.
- Ashida, H., Mimuro, H. and Sasakawa, C. (2015). Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol 6: 219.
- Bernardini, M. L., Mounier, J., d'Hauteville, H., Coquis-Rondon, M. and Sansonetti, P. J. (1989). Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci U S A 86(10): 3867-3871.
- Cappuccino, J. G. and Welsh, C.T (2017). Microbiology: A Laboratory Manual, 11th Edition. ISBN: 978-0134298672.
- Chantret, I., Rodolosse, A., Barbat, A., Dussaulx, E., Brot-Laroche, E., Zweibaum, A. and Rousset, M. (1994). Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: evidence for glucose-dependent negative regulation. J Cell Sci 107 (Pt 1): 213-225.
- Freshney, R. I. (2010). Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications. Sixth Edition. ISBN: 9780470528129.
- Koestler, B. J., Fisher, C. R. and Payne, S. M. (2018). Formate promotes Shigella intercellular spread and virulence gene expression. MBio 9(5): e01777-18.
- Kotloff, K. L., Riddle, M. S., Platts-Mills, J. A., Pavlinac, P. and Zaidi, A. K. M. (2018). Shigellosis. Lancet 391(10122): 801-812.
- Makino, S., Sasakawa, C., Kamata, K., Kurata, T. and Yoshikawa, M. (1986). A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell 46(4): 551-555.
- Oaks, E. V., Wingfield, M. E. and Formal, S. B. (1985). Plaque formation by virulent Shigella flexneri. Infect Immun 48(1): 124-129.
- Puhar, A. and Sansonetti, P. J. (2014). Type III secretion system. Curr Biol 24(17): R784-791.
- Puhar, A., Tronchère, H., Payrastre, B., Nhieu, G. T. and Sansonetti, P. J. (2013). A Shigella effector dampens inflammation by regulating epithelial release of danger signal ATP through production of the lipid mediator PtdIns5P. Immunity 39(6): 1121-1131.
- Sansonetti, P. J., Kopecko, D. J. and Formal, S. B. (1982). Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 35(3): 852-860.
- Schroeder, G. N. and Hilbi, H. (2008). Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21(1): 134-156.
- Shigellosis-Chapter 3-2018 Yellow Book | Travelers’ Health | CDC. Available at: https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/shigellosis.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sharma, A. and Puhar, A. (2019). Plaque Assay to Determine Invasion and Intercellular Dissemination of Shigella flexneri in TC7 Human Intestinal Epithelial Cells. Bio-protocol 9(13): e3293. DOI: 10.21769/BioProtoc.3293.
Category
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell staining > Whole cell
Cell Biology > Cell viability > Cell death
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











