- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detachment Procedure of Bacteria from Atmospheric Particles for Flow-cytometry Counting
(*contributed equally to this work) Published: Vol 9, Iss 12, Jun 20, 2019 DOI: 10.21769/BioProtoc.3273 Views: 5583
Reviewed by: Alexandros AlexandratosVemika ChandraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assay for Assessing Mucin Binding to Bacteria and Bacterial Proteins
Lubov S. Grigoryeva [...] Nicholas P. Cianciotto
Mar 5, 2021 4766 Views

Flow Cytometry-based Measurement of Reactive Oxygen Species in Cyanobacteria
Soumila Mondal and Shailendra P. Singh
May 20, 2022 3835 Views

Flow Cytometric Quantification of HIV-1-Infected Cells Expressing Either Abortive or Elongated HIV-1 Transcripts Using Flow-FISH
Shirley Man [...] Neeltje A. Kootstra
Jul 20, 2025 2156 Views
Abstract
The protocol separates bacteria from atmospheric particles, obtaining with greater precision their abundance in the atmospheric deposition. This procedure is similar to the one used to separate bacteria in streambed sediments. The detachment procedure consists of a chemical treatment with sodium pyrophosphate and Tween 20 and a physical treatment with agitation and ultrasonic bath to disperse the bacteria in the liquid sample. We recover the total (free and attached) bacteria by generating a density gradient with Nycodenz by centrifugation. The techniques prior to this procedure do not include the microorganisms that are attached to the aerosol particles and, therefore, considerably underestimate the total load and deposition of airborne microorganisms.
Keywords: Airborne bacteriaBackground
The study of airborne microorganisms (bioaerosol) is relevant to understand their long-range dispersal and to assess their meteorological consequences (Kellogg and Griffin 2006; Creamean et al., 2013). Airborne microorganisms, aerosolized from soil or sea surface, can be transported large distances by the global atmospheric circulation expanding their distribution ranges (Aller et al., 2005; Hervàs et al., 2009; Yamaguchi et al., 2012). They are removed from the atmosphere by wet (rain/snow) and dry deposition (sedimentation), affecting the microbial abundance, composition, and distribution in recipient ecosystems (Peter et al., 2014; Reche et al., 2018). In addition, bacteria in the atmosphere can act as condensation nuclei of clouds and ice affecting meteorological and climatic conditions (Fröhlich-Nowoisky et al., 2016). Viruses and bacteria usually are air-dispersed attached to soil-dust or to marine organic aggregates, not as free cells or particles (Yamaguchi et al., 2012; Reche et al., 2018). This protocol allows detaching bacteria and viruses from atmospheric particles to quantify them more accurately using flow cytometry (Reche et al., 2018). This protocol can be relevant in the fields of microbial ecology, aerobiology, and atmospheric sciences. This procedure was inspired in another similar by Amalfitano and Fazi (2008) to detach benthic bacteria from stream sediments.
The detachment procedure consists of two steps: first, a chemical treatment by adding sodium pyrophosphate and Tween 20 to break the links to particles and aggregates; and a second physical treatment by shaking and incubation in an ultrasonic bath to disperse the bacteria into the liquid phase. To recover all the bacteria, we generate a density gradient with Nycodenz by centrifugation obtaining four layers (from top to bottom: supernatant, microbial layer, Nycodenz and pellet). We retrieve completely the layer with microorganisms that were suspended in sterilized Milli-Q water up to the original volume, fixed and stored at -80 °C until the flow cytometry analysis.
Materials and Reagents
- Sterile universal pipette tips (Scharlab, S.L., catalog numbers: 000P0210-1 [0.5 μl-10 μl]; 00PC5200-1 [10-100 μl]; 000P1000-1 [100-1,000 μl])
- 1.5 ml microcentrifuge tubes (Labbox Labware S.L., catalog number: PCRP-015-500)
- Flow cytometry tubes (Sarstedt, catalog number: 55.1579)
- Syringe 0.2 μm-filter (Sartorius, MinisartR, catalog number: 16534-k)
- Syringe and needle (ICOplus3, Novico, catalog number: 164624.8)
- Simport Cryovial (SimportTM Scientific, catalog number: T310-5A)
- Bemis Parafilm M (Bemis, catalog number: PM996)
- Polycarbonate membrane filter
- Sodium pyrophosphate tetrabasic (Na4P2O7) (Sigma-Aldrich, catalog number: P8010)
- Tween 20, Polyoxyethylenesorbitan monolaurate (C26H50O10) (Sigma-Aldrich, catalog number: P1379)
- Nycodenz 60% (w/v) (C19H26I3N3O9) (NycoPrepTM Universal, Axis-Shield, catalog number: 31000.01)
- Milli-Q water
- Paraformaldehyde and glutaraldehyde
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148-500G)
- Glutaraldehyde 25% (Scharlau, catalog number: GL01701000)
- Phosphate-buffered saline tablets (Sigma-Aldrich, catalog number: P4417)
- Sheath fluid (BD FACSFlow, catalog number:342003)
- SYBR Green I (Sigma-Aldrich, catalog number: S9430)
- Dimethyl sulfoxide (DMSO) (C2H5OS) (Sigma-Aldrich, catalog number: 472301)
- Latex 1 μm-beads (Polysciences, catalog number: 18860-1)
- Phosphate-buffered saline (PBS) solution
- Sodium pyrophosphate tetrabasic stock solution (see Recipes)
- Paraformaldehyde and glutaraldehyde solution (see Recipes)
- SYBR Green I solution (see Recipes)
Equipment
- Eppendorf Pipettes (0.5-10 μl, 10-100 μl, 100-1,000 μl)
- Glass beaker
- WiseStir Hotplate stirrer (MSH-20A)
- Vortex mixer for detachment procedure (Velp scientifica, catalog number: F202A0173)
- Ultrasonic cleaner for detachment procedure (P. Selecta, Ultrasons)
- Refrigerated centrifuge (Eppendorf Microcentrifuge, model: 5415R)
- Light source (lamp)
- Ultrasonic cleaner for flow cytometry (Bransonic, model: 200)
- Vortex mixer for flow cytometry (VWR International)
- Flow cytometer (Becton, Dickinson and Company, BC Biosciences, BD FACSCaliburTM)
- Freezer -80 °C (Thermo ScientificTM, HerafreezeTM, model: SAT 901120200, catalog number: HFU686BA)
Software
- BD CellQuestTM Pro Software (Becton Dickinson and Company, https://www.bd.com/en-uk/products/molecular-diagnostics/cytometric-analysis-products)
Procedure
This procedure detaches bacteria and viruses from aerosol (dust and organic aggregates) particles for flow-cytometry counting (Reche et al., 2018). It is similar to the procedure used for benthic bacteria attached to streambed sediments by Amalfitano and Fazi (2008). After this detachment procedure, bacteria are unattached, dispersed in sterile MilliQ-water and they, then, can be easily counted by flow cytometry representing the total of bacteria associated to atmospheric particles. The atmospheric samples come from dry and wet collectors. We take 3 ml-samples into 5 ml-criovials, fix them with paraformaldehyde and glutaraldehyde (10% + 0.05% final concentration) in the field, freeze them in liquid nitrogen, and keep them at -80 °C until analysis. Dry deposition is recovered from the collector using sterile Milli-Q water. More details on dry and wet collectors can be found in Reche et al. (2018).
Here, we mainly expose the steps for the bacterial counting (Gasol and Del Giorgio, 2000), but the previous steps on the retrieve of the microbial layer are also common for viruses.
- Bacteria detachment procedure
- Take 718 μl of atmospheric deposition samples in 1.5 ml-microcentrifuge tubes. This aliquot contains both free and attached to particles bacteria.
- Add 56.4 μl of the working solution (see Recipes) of the chelating agent sodium pyrophosphate (ca. 0.1% final concentration), and 7.5 μl of the detergent Tween 20 (0.5% final concentration) to reduce the adhesion of bacteria to the mineral and organic particles of the aerosol (Figure 1A).

Figure 1. Scheme of the different steps for the detachment procedure. A. Chemical treatment using the sodium pyrophosphate as chelator and Tween 20 as detergent. B. Physical treatment shaking the samples and ultrasonic bath. C-D. Purification of the microbial layer using Nycodenz to generate a density gradient of four layers. E-F. Select the microbial layer and suspend it in sterilized Milli-Q water up to the initial sample volume in a cryovial. - Shake the samples for 30 min at ~720 rpm in a vortex (Figure 1B), and place them in an ultrasonic bath for 1 min to disperse the attached bacteria into the liquid phase. It is important not to place the samples in the ultrasonic bath for more than 1 min to prevent cellular damages during the detachment procedure.
- Place carefully approx. 718 μl of Nycodenz beneath of treated sample using a needle long enough to reach the bottom of a 1.5 ml-microcentrifuge tube (Figure 1C). The syringe with the Nycodenz should not disturb the content of the microcentrifuge tube or has air bubbles. This is important to form properly the density gradient.
- Centrifuge the microcentrifuge tubes at 14,000 x g for 90 min at 4 °C (Figure 1D) in a density gradient using Nycodenz as medium to separate four density layers (top-down): supernatant, microbial layer, Nycodenz and pellet (Figure 1E). The pellet is not always visible. The observation of the pellet depends on particle loading in the atmospheric deposition and it is mainly observed when atmospheric deposition contains Saharan dust.

Figure 2. Backlight picture of a microcentrifuge tube after microcentrifugation (after the Step 5). After the Step 5, the microcentrifuge is with four layers (top-down): supernatant, microbial layer, Nycodenz and dust pellet. - Identify the microbial layer with a backlight (Figure 2, Video 1) and carefully aspire completely this layer using a tip with the pipette (Figure 1E, Video 2).
- Place the microbial layer (200 μl) into cryovial tubes and suspend it in 518 μl of sterilized Milli-Q water to reach the initial volume (718 μl) (Figure 1F). Then, fix the sample again with paraformaldehyde and glutaraldehyde (final concentration 1% + 0.05%) and store it at -80 °C until the flow cytometry analysis.Video 1. Backlight video moving the microcentrifuge tube to clearly identify the contrasted, milky structure of the microbial layerVideo 2. Backlight video to show the procedure to retrieve the microbial layer
- Bacterial counting by flow cytometry
- Once the cytometer is clean, calibrate the flow of the cytometer each working day. First, weigh a tube containing Milli-Q water, this value is the initial weight (Winitial) in mg (= μl), then run this Milli-Q water sample for 10 min at low flow in the cytometer, and re-weigh the tube again, this value is the final weight (Wfinal) in mg (= μl). Second, estimate the working flow using the next equation. This working flow usually ranges from 33.6 μl min-1 to 42.2 μl min-1.
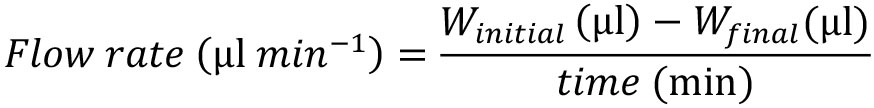
- Set the flow cytometer with excitation at 448 nm, the fluorescence Side Scatter (SSC) and Forward Scatter (FSC) parameters in logarithmic scale, the threshold on green fluorescence (FL1-H) and then run a Milli-Q water sample. Keep the events at very low level (no green fluorescence must be detected).
- Defrost the samples and prepare 500 μl-aliquots by triplicate in flow cytometer tubes, add 5 μl of SYBR Green I (1:1,000) to stain the bacteria, vortex the cytometer tubes and incubate them in the dark for 10 min at 25 °C, add 10 μl of the suspension of fluorescent latex beads (concentration 1 x 105) as an internal reference, and then, run the samples at low flow rate. First, run a Milli-Q water sample as negative control and, then, the experimental samples until around 10000 events were acquired. Events have to range between 200 and 800 per second to avoid coincidence. The samples have to be diluted when events are higher than 800 per second.
- Save the data for further processing with the software BD CellQuest Pro.
- Once the cytometer is clean, calibrate the flow of the cytometer each working day. First, weigh a tube containing Milli-Q water, this value is the initial weight (Winitial) in mg (= μl), then run this Milli-Q water sample for 10 min at low flow in the cytometer, and re-weigh the tube again, this value is the final weight (Wfinal) in mg (= μl). Second, estimate the working flow using the next equation. This working flow usually ranges from 33.6 μl min-1 to 42.2 μl min-1.
Data analysis
Data analysis of cytograms is performed using BD CellQuest Pro software. Bacterial cells are identified using their signature in bivariate plots (green fluorescence FL1-H vs. side light scattered SSC), and the preselected gates that discriminate bacterial position and size. The success of the detachment procedure is routinely controlled. The same samples are counted without the detachment procedure (Figures 3A and 3C) and after this procedure (Figures 3B and 3D). Samples submitted to the detachment procedure represent total (free and atmospheric particle-attached) bacteria. 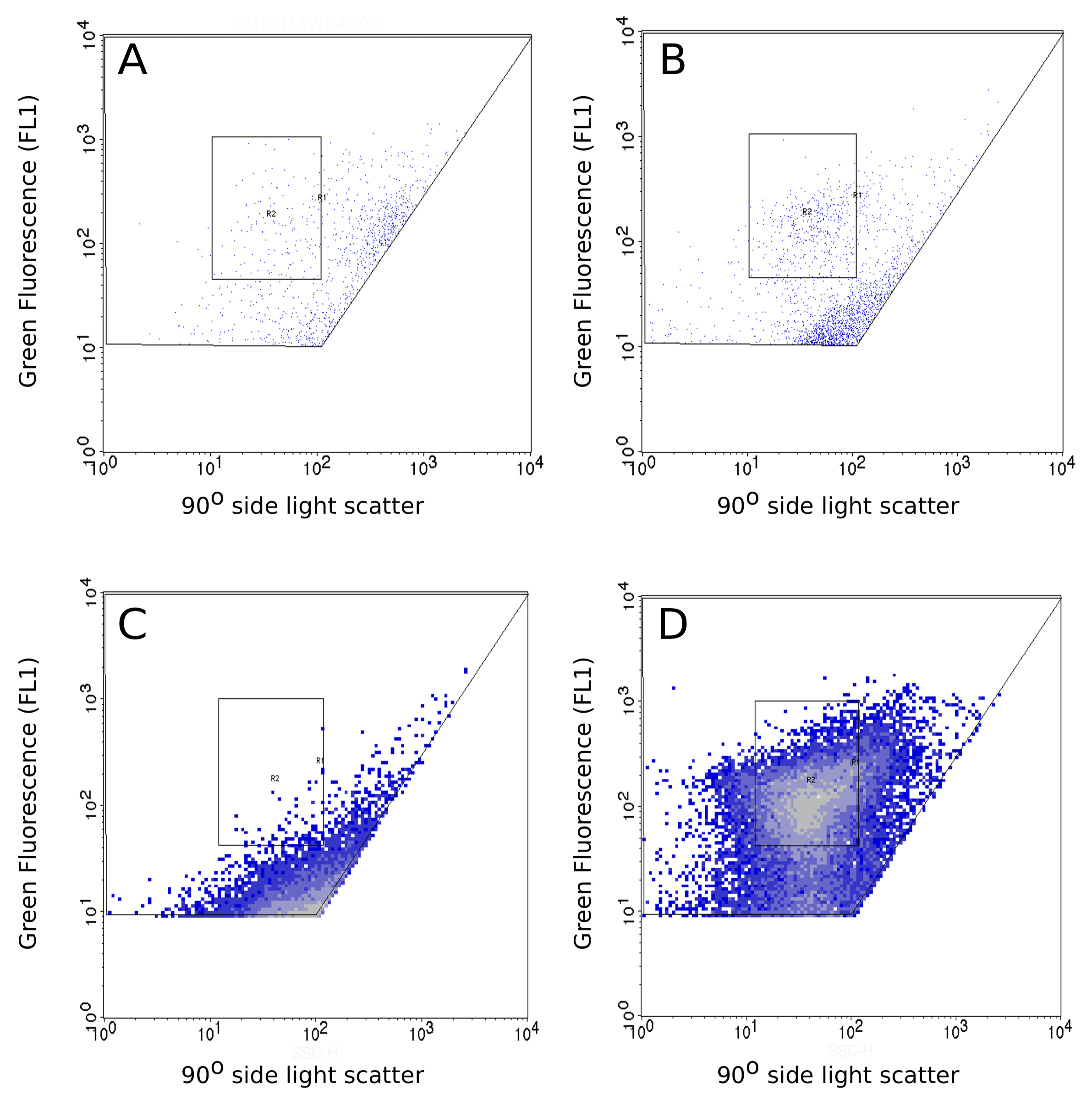
Figure 3. Comparison between cytograms of the same sample without the detachment treatment and after undergoing this treatment. A-B. Cytograms of a wet deposition sample under a meteorological scenario of Atlantic influence without the detachment procedure (A) and after being submitted to the detachment procedure (B). C-D. Cytograms of a wet deposition sample under a meteorological scenario of Saharan dust intrusion without the detachment procedure (C) and after being submitted to the detachment procedure (D). The x-axis corresponds to side light scatter (SSC) and y-axis corresponds to green fluorescence (FL1-H). The R2 rectangle is the selected area for the bacterial counting. Events outside this area are impurities.
Notes
- Nycodenz has to be stored at or below 20 °C and protected of the light.
- Latex beads have to be stored at -20 °C and the bead suspension has to be incubated in an ultrasonic bath for 10 min before the counting with the flow cytometer.
Recipes
- Sodium pyrophosphate tetrabasic
Preparation of stock solution (1 M):- Weigh 26.59 g of sodium pyrophosphate tetrabasic and dilute it in 100 ml sterile Milli-Q water
- Prepare the working solution (0.1 M) taking a 10 ml of the stock solution and dilute it in 100 ml of sterile Milli-Q water
- Paraformaldehyde and glutaraldehyde solution (final concentration 1% + 0.05%)
- Add 100 g of paraformaldehyde (Sigma, P6148) to 880 ml of Milli-Q water in a glass beaker and cover it with Parafilm
- Dissolve the paraformaldehyde using a magnetic stirrer on a heating plate at 90 °C for a period of 24 h to 72 h under a fume hood. The water must be close to the boiling point
- Add 100 ml of phosphate-buffered saline (PBS) solution. This solution is previously prepared by dissolving a tablet of PBS (Sigma, P4417) in 200 ml Milli-Q water.
- Add 20 ml of the glutaraldehyde solution 25%
- Filter through a polycarbonate membrane filter (25 mm diameter 0.2 μm), take aliquots of 5-20 ml in cryovials and store them at -80 °C (long term storage) or at -20 °C (working solution)
- SYBR Green I solution (1:1,000)
Dilute SYBR Green I with DMSO to 1:1,000 and store at -20 °C protected of the light
Acknowledgments
This work was supported by the Spanish Ministry of Economy and Competitiveness (project CGL2015-73250-JIN) and an MSc scholarship for research initiation of the Vicerectorate for Research and Transfer of the University of Granada to CMA. We also thank Dr. Gustavo Ortiz Ferrón for his guidance in flow cytometry. This procedure was previously used in Reche et al. (2018) and is an adaptation for atmospheric particles of the procedure by Amalfitano and Fazi (2008) to streamed bed sediments.
Competing interests
The authors declare no competing interests.
References
- Aller, J. Y., Kuznetsova, M. R., Jahns, C. J. and Kemp, P. F. (2005). The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J Aerosol Sci 36(5-6): 801-812.
- Amalfitano, S. and Fazi, S. (2008). Recovery and quantification of bacterial cells associated with streambed sediments. J Microbiol Methods 75(2): 237-243.
- Creamean, J. M., Suski, K. J., Rosenfeld, D., Cazorla, A., DeMott, P. J., Sullivan, R. C., White, A. B., Ralph, F. M., Minnis, P., Comstock, J. M., Tomlinson, J. M. and Prather, K. A. (2013). Dust and biological aerosols from the Sahara and Asia influence precipitation in the western U.S. Science 339(6127): 1572-1578.
- Fröhlich-Nowoisky, J., Kampf, C. J., Weber, B., Huffman, J. A., Pöhlker, C., Andreae, M. O., Lang-Yona, N., Burrows, S. M., Gunthe, S. S., Elbert, W., Su, H., Hoor, P., Thines, E., Hoffmann, T., Després, V. R. and Pöschla, U. (2016). Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmos Res 182: 346-376.
- Gasol, J. M., and Del Giorgio, P. A. (2000). Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar 64(2): 197-224.
- Hervàs, A., Camarero, L., Reche, I., and Casamayor, E. O. (2009). Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol 11(6): 1612-1623.
- Kellogg, C. A. and Griffin, D. W. (2006). Aerobiology and the global transport of desert dust. Trends Ecol Evol 21(11): 638-644.
- Peter, H., Hortnagl, P., Reche, I. and Sommaruga, R. (2014). Bacterial diversity and composition during rain events with and without Saharan dust influence reaching a high mountain lake in the Alps. Environ Microbiol Rep 6(6): 618-624.
- Reche, I., D'Orta, G., Mladenov, N., Winget, D. M. and Suttle, C. A. (2018). Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J 12(4): 1154-1162.
- Yamaguchi, N., Ichijo, T., Sakotani, A., Baba, T. and Nasu, M. (2012). Global dispersion of bacterial cells on Asian dust. Sci Rep 2: 525.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Araya, C. M., Cazorla, A. and Reche, I. (2019). Detachment Procedure of Bacteria from Atmospheric Particles for Flow-cytometry Counting. Bio-protocol 9(12): e3273. DOI: 10.21769/BioProtoc.3273.
Category
Environmental science > Bacterium > Isolation
Microbiology > Microbial cell biology > Cell isolation and culture
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











