- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorescence HPLC Analysis of the in-vivo Activity of Glucosylceramide Synthase
Published: Vol 9, Iss 12, Jun 20, 2019 DOI: 10.21769/BioProtoc.3269 Views: 6637
Reviewed by: Neelanjan BoseIstvan StadlerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of the Activity of Wildtype and Disease-Causing ALPK1 Mutants in Transfected Cells With a 96-Well Format NF-κB/AP-1 Reporter Assay
Tom Snelling
Nov 20, 2024 1602 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2277 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 481 Views
Abstract
Almost all functions of cells or organs rely on the activities of cellular enzymes. Indeed, the in-vivo activities that directly represent the cellular effects of enzymes in live organs are critical importance to appreciate the roles enzymes play in modulating physiological or pathological processes, although assessments of such in-vivo enzyme activity are more difficult than typical test-tube assays. Recently, we, for the first time, developed a direct and easy-handling method for HPLC analyzing the in-vivo activity of glucosylceramide synthase (GCS). GCS that converts ceramide into glucosylceramide is a limiting-enzyme in the syntheses of glycosphingolipids and is one cause of cancer drug resistance. In our method developed, rubusoside nanomicelles delivers fluorescence N-[6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-sphingosine (NBD C6-ceramide) into mice, tissues uptake the cell-permeable substrate, and GCS converts it into NBD C6-glucosylceramide in all organs simultaneously. Further, HPLC analyzes the extracted NBD C6-glucosylceramide to assess alterations of the in-vivo GCS activities in tissues. This method can be broadly used to assess the in-vivo GCS activities in any kind of animal models to appreciate either the role GCS plays in diseases or the therapeutic efficacies of GCS inhibitors.
Keywords: In-vivo enzyme activityBackground
Glucosylceramide synthase (GCS; EC22.41.80; also known as ceramide glucosyltransferase) catalyzes ceramide glycosylation, transferring glucose residues from uridine diphosphate-glucose (UDP-glucose) to ceramide (Cer) and thereby producing glucosylceramide (GlcCer) (Basu et al., 1968; Ichikawa et al., 1996). Cer glycosylation by GCS is the first and rate-limiting step for the synthesis of glycosphingolipids (GSLs), over 400 species in human (Yu et al., 2009; Merrill, 2011; D'Angelo et al., 2013). Highly localized on GSL-enriched microdomains (GEMs) of cellular membranes, GSLs play crucial roles in modulating membrane integrity and functions, including cell-cell recognition, interaction and communication (Fishman and Brady, 1976; Yoshizaki et al., 2008; D'Angelo et al., 2013). GSLs are involved in mediating cell proliferation, differentiation, immuno-response and oncogenic transformation (Hakomori, 1981; Kasahara and Sanai, 2000; Hakomori, 2002).
Aberrant GCS or GSLs highly associate with several diseases. Recent studies concordantly indicate that enhanced GCS is one cause of cancer drug resistance (Liu et al., 2001, 2008, 2010 and 2013; Chiu et al., 2015; Roh et al., 2015). Further, inhibition of GCS, thereby decreasing GlcCer accumulation, is a treatment option for Gaucher’s disease (McEachern et al., 2007) and type II diabetes (Aerts et al., 2007; Zhao et al., 2007).
Quantitative assessment of GCS activity is essential for evaluating the roles Cer glycosylation playing in cell functions, as well as in therapeutic efficacies of relevant disease treatments. After Basu’s work (Basu et al., 1968), several additional methods have been developed to analyze GCS activities (Basu et al., 1987; Shukla and Radin, 1990; Ichikawa et al., 1996; Hayashi et al., 2005). Besides UDP-[3H]glucose being incorporated into ceramide to generate [3H]GlcCer (Shukla and Radin, 1990; Lavie et al., 1997; Liu et al., 1999), N-[6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-sphingosine (NBD C6-Cer) has been used as an acceptor to produce NBD C6-GlcCer by the prepared GCS protein in optimal in-vitro conditions (Ichikawa et al., 1996; Bourteele et al., 1998; Hayashi et al., 2005). The HPLC method based on NBD C6-Cer has proven to be a highly sensitive and reproducible assay for assessing GCS activity in-vitro assays (Ichikawa et al., 1996; Hayashi et al., 2005). Convergently, previous studies have shown that NBD C6-Cer can be used as an exogenous substrate for characterizing cellular Cer glycosylation and assessing GCS activities with thin-layer chromatography (TLC) and spectrometry (Gupta et al., 2010; Liu et al., 2010; Gupta et al., 2012). With nanoparticle-based delivery of NBD C6-Cer, we developed a rapid, efficient, and fully quantitative HPLC analysis for assessing GCS in-vivo activity in live mice (Khiste et al., 2017).
Rubusoside (RUB) has been used to improve solubility and consequently bioavailability of etoposide (Zhang et al., 2012). RUB based nanomicelles (Cer-RUB) significantly increases the aqueous solubility and bioavailability of NBD C6-Cer in vivo (Khiste et al., 2017). After intraperitoneal administration, almost all tissues efficiently uptake and use NBD C6-Cer as substrate for Cer glycosylation by GCS. Analysis of the extracted sphingolipids with HPLC allows us directly assess the in-vivo activities that GCS simultaneously conduct among tissues in live mice. This direct, specific and effective approach can be applied to measure intracellular and intra-organ GCS activities in live cells and tissues, enabling further assessments of the roles played by GCS in diseases. Notably, this methodology may have considerable value in predicting and monitoring therapeutic efficacy of various anticancer drugs, and of other therapeutic agents used in the treatments of GCS-related diseases.
Materials and Reagents
- Falcon 100 mm cell culture dish (Corning, catalog number: 353003)
- Dishes 35-mm (Corning, catalog number: 353001)
- Paper towels Scott C-fold (Uline, catalog number: 560801)
- BD 1 ml insulin syringes (Fisher Scientific, catalog number: 22-253-260)
- Pasteur pipettes (Fisher Scientific, catalog number: 13-678-20D)
- Microcentrifuge tubes, 1.5 ml (Fisher Scientific, catalog number: 0548129)
- Polypropylene centrifuge tubes, 15 ml (Fisher Scientific, catalog number: 22-010-076)
- Polypropylene centrifuge tubes, 50 ml (Fisher Scientific, catalog number: 05-539-6)
- Screw thread vials, glass 4 ml, amber (Thermo Fisher Scientific, catalog number: C4015-2W)
- Screw thread vials, glass 4 ml, transparent (Thermo Fisher Scientific, catalog number: C4015-21)
- LVITM (low volume insert) vials, 0.3 ml, amber glass (Wheaton, catalog number: 225328)
- 96-well black flat bottom polystyrene microplates for fluorescence measurement (Fisher Scientific, catalog number: 07-200-589)
- Polystyrene disposable pipets, 10 ml, sterile (Fisher Scientific, catalog number: NC9868325)
- Glass pipettes, 1 ml, sterile (Fisher Scientific, catalog number:12-724)
- Pipet tips, sterile, 200 μl and 1 ml (Fisher Scientific, catalog numbers: 02-707-502, 02-707-509)
- 0.45-μm nylon filter (Thermo Fisher Scientific, catalog number: 151-4045)
- Sterile alcohol prep pads (Dynarex, catalog number: 1114)
- Athymic nude mice (Foxn1nu/Foxn1+, 4-5 weeks, female from ENVOGO, catalog number: 069(nu)/070(nu/+))
- Human ovary cancer OVCAR-3 (ATCC, catalog number: ATCC® HTB-161TM) (Khiste et al., 2017)
- Mice carrying OVCAR-3 tumor xenografts and controls
- Inoculate OVCAR-3 cells (purchased from ATCC; ~3-5 passages, 1 x 106/20 μl per mouse) in serum-free medium subcutaneously inoculate into the left flanks of athymic nude mice (Foxn1nu/Foxn1+, 4-5 weeks, female; Harlan).
- In the control group, we only inoculate 20 μl serum-free medium. Once tumors sizes reach ~0.6 cm in diameters (~24 days), we will inject NBD Cer-RUB nanomicelles intraperitoneally in these mice for ceramide glycosylation.
- RPMI 1640 Medium (Thermo Fisher Scientific, catalog number: 11875-093), store at 4 °C
- Fetal bovine serum (FBS), heat inactivated (Thermo Fisher Scientific, catalog number: 10082147), store at -20 °C
- Phosphate buffer saline (PBS, 10x) (Thermo Fisher Scientific, catalog number: 70011-044), keeping at 4 °C for using
- NBD C6-ceramide (N-[6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]hexanol]-D-erythro-sphingosine) (Avanti Polar Lipids, catalog number: 810209), store at -20 °C
- NBD C6-glucosylceramide, (N-hexanoyl-NBD-glucosylceramide) (Matreya, catalog number: 1622-001)
- PierceTM BCA protein assay kit (Thermo Fisher Scientific, catalog number: 23227)
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: 9048-46-8), store at 4 °C
- NP40 Cell Lysis Buffer (Thermo Fisher Scientific, catalog number: FNN0021), store at -20 °C
- ProteaseArrestTM for Mammalian (G-Bioscience, catalog number: 786-433), store at 4 °C
- Methanol for HPLC, ≥ 99.9% (Sigma-Aldrich, catalog number: 67-56-1)
- Chloroform for HPLC, ≥ 99.5% (Sigma-Aldrich, catalog number: 67-66-3)
- Water for HPLC (Fisher Chemical, catalog number: 7732-18-5)
- Ortho-phosphoric acid for HPLC (Sigma-Aldrich, catalog number: 7664-38-2)
- Ethanol, anhydrous (Fisher Chemical, catalog number: A405-20)
- Rubusoside (RUB) (Sigma-Aldrich, catalog number: 62933-10MG)
- NBD C6-ceramide-rubusoside (NBD Cer-RUB) solution (see Recipes)
- NBD Cer-RUB nanomicelles (see Recipes)
- NP40 protein lysis solution (see Recipes)
- Solvent system for HPLC analysis (see Recipes)
Solvent A
Solvent B - NBD C6-ceramide or NBD C6-glucosylceramide standard solutions (see Recipes)
10 μM NBD C6-ceramide
10 μM NDB C6-glucosylceramide
Note: Store or keep all reagents at room temperature, except indicated items.
Equipment
- Pipette aids and pipette
- High precision microdissection scissors (Fisher Scientific, catalog number: 08-953-1B)
- Analytical balance (Mettler Toledo, catalog number: ME54TE/00)
- Freezer, -20 °C (Fisher Scientific, catalog number: 18LC-16WW-FS)
- Freezer, -80 °C (Thermo Scientific, catalog number: ULT1386-5-D41)
- Standard cell culture hood
- Vortex mixers (Fisher Scientific, catalog number: 02-215-360)
- HPLC system, Agilent 1220 Infinity II LC with Agilent 1260 fluorescence detector (Agilent Technologies)
- HPLC Colum, ZORBAX Rx-SIL 4.6 x 250 mm, 5 μM (Agilent Technologies, catalog number: 880975-901)
- Centrifuge (Eppendorf, model: 5804 R)
- Centrifuge (Eppendorf, model: 5424 R)
- Water bath and incubator
- Reacti-Vap evaporators (Thermo Fisher Scientific, catalog number: TS-18826)
- Ultrasonic processor (Fisher Scientific, model: CPX130PB)
- Microplate Reader (BioTek, Synergy HTX multi-mode reader)
Software
- Prism 5v for statistical analysis (GraphPad Software Inc.)
- Agilent OpenLab CDS (EZChrom edition, version A.04.05) for HPLC analysis
Procedure
Briefly, we inject NBD C6-Cer-RUB nanomicelles (Zhang et al., 2012; Hosain et al., 2016; Khiste et al., 2017) into tumor-bearing mice, and the live tissues of mice convert NBD C6-ceramide into NBD C6-glucosylceramide. Three hours after injection, we collect all tissues, including tumors, and then extract the lipids. Further, we apply HPLC to analyze NBD glucosylceramide and assess the in vivo GCS activities of tissues.
- Injection of NBD Cer-RUB into mice and in vivo glycosylation
- Prepare NBD Cer-RUB solution (NBD C6-Cer 0.25 mg/ml) in sterile condition (Recipe 1) and keep on ice (Figure 1A).
- Weigh body weight of each mouse on balance and calculate the injection volume of NBD C6 Cer-RUB solution, based on 1 mg/kg body weight for each mouse bearing OVCAR-3 tumor xenografts (Materials and Reagents #20). For instance, we inject 100 μl NBD C6 Cer-RUB solution containing 25 μg NBD C6-ceramide to a 25 g mouse.
- In animal operating hood, take NBD Cer-RUB solution into a U-100 insulin syringe, inject intraperitoneally into mice (Recipes 1, 5 cases/group) following wipe inject-site skin with 70% alcohol (Figure 1B).
- Keep injected mice in cages for 3 h, allowing uptake of NBD C6-ceramide and ceramide glycosylation in live animals. There are approximately 10 min intervals among the injections.
- Euthanize mouse with cervical dislocation and dissect tissues, including liver, lungs and tumors from three mice per group, according to the injection order.
- Rinse tissues with sterile PBS in 35-mm dishes twice and absorb with sterile paper towels.
- Dissect and put tissue of each mouse into microcentrifuge tubes, keep on ice or store at -80 °C.
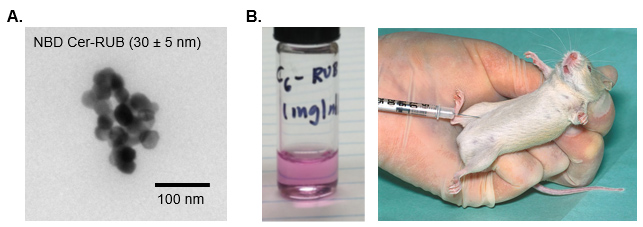
Figure 1. Intraperitoneal injection of NBD Cer-RUB nanomicelle solution to mice. A. NBD Cer-RUB complex are nanomicelles. B. NBD Cer-RUB is solubilized in RPMI-1640 medium and intraperitoneally injected into mouse for the in vivo glycosylation by GCS.
- Tissue collection and extraction of lipids and proteins
- Dissect and weigh two of 50 mg pieces from the same tissue of all samples (liver, lung, tumor from five mice per group), and put in 2 microcentrifuge tubes containing 50 μl sterile PBS, respectively. Use one set of samples for lipid extraction and another one for protein extraction.
- Mince tissue with micro-scissor and sonicate it with an ultrasonic processor in PBS on ice.
- For lipid extraction:
- Add 100 μl water and transfer the tissue suspension into a 4-ml transparent glass vial for all samples.
- Add 0.85 ml of water, 1.0 ml methanol and 1.0 ml chloroform in the glass vial and vortex for 30 s to mix. Keep samples at room temperature for 20 min and vortex for 30 s after every 5 min.
- Centrifuge the glass vial tube at 4,500 x g, room temperature for 10 min, to separate the organic and aqueous phases.
- Gently hold glass vial and transfer of the organic phase (bottom layer) with Pasteur pipette into a new amber 4-ml glass vial (Figure 2, Video 1).
- Take 5 μl organic solution and add into a fluorescence 96-well plate with 145 μl methanol in duplicate. Measure fluorescence units in a microplate reader (BioTek Synergy HT, λ excitation 470 nm, λ emission 530 nm). The average fluorescence unites will be used to normalize all samples.
- Transfer 800 μl organic solution of each sample with glass pipette into an amber 4-ml glass vial, and dry with nitrogen gas completely (~20 min). Tightly close the vials and store all samples at -20 °C for further use.

Figure 2. Transferring lipid extracts after extraction. After centrifugation, the organic solution is in the lower phase of vials. Transferring the lower phase, which contains extracted fluorescence lipids with Pasteur pipette into a new amber glass vial. This step needs to practice before experiments.Video 1. Transferring lipid extracts in the lower-phase with Pasteur pipette. Holding vials between left thumb and index fingers with ~65°, insert pipette tip into the bottom layer to the lowest corner of the sample vial. Slowly aspirate the lower-phase solution only (~900 µl) with pipette-aid, and then transfer it into the new glass vial.
- For protein extraction:
- Add 200 μl NP40 lysis buffer with ProteaseArrest (Recipe 3) and mix with tissue suspension (from Steps B1 and B2), and vortex for 30 s.
- Incubate the tubes on ice for 40 min, with vortex for 30 s per 5 min.
- Centrifuge samples at 9,400 x g, 4 °C for 10 min.
- Collect the supernatant of each sample in new microcentrifuge tubes, and store at -80 °C, except the portion for further protein measurement.
- Add 2 μl of protein extracts in triplicate and mix with 100 μl BCA reagents (50:1, Reagent A:B), into 96-well plates. Use bovine serum albumin (BSA, 0, 5, 10, 20, 40 μg) to generate the standard curve.
- Measure absorbance in a microplate reader (λ = 562 nm; BioTek Synergy HT) and obtain protein concentrations of samples (x μg/μl) with the BSA standard. The total protein amount: Pro = x × 160 (μg) for each sample (from 50 mg tissue used).
- HPLC Analysis of NBD C6-Glucosylceramide of mouse tissues
- Characterize and measure NBD C6-glucosylceramide with NBD C6-ceramide in standard solutions.
- Connect the Agilent 1220 Infinity LC system with column (ZORBAX Rx-SIL 5 μm, 4.6 x 250 mm, normal phase) and solvents, and equilibrate with Solvent A, at flow rate 1 ml/min for 60 min.
- Set up OpenLab CDS EZChrom program: flow rate 1 ml/min, injection volume 5 μl, linear gradient for 14 min (Solvent A 100%~0, and Solvent B 0~100%), and fluorescence detection (λ excitation 470 nm, λ emission 530 nm).
- Add 100 μl solutions of NBD C6-ceramide (Cer) and NBD C6-glucosylcerade (GlcCer) into the LVI vials, respectively. These solutions contain both Cer and GlcCer at increasing concentrations (0, 0.032, 0.063, 0.125, 0.25, 0.5 and 1 μM; see Recipe 5) (Figure 1A).
- Load the LVI vials in the carousel of HPLC system, run analysis triplicate and save the files.
- Characterize NBD glucosylceramide generated of samples.
- Add 100 μl of methanol/chloroform into in the amber glass vial of liver extracts to reconstitute lipids (Step B4f).
- Transfer 50 μl of reconstituted liver lipids (~2,000 fluorescence units) into the LVI vials.
- Add 2 μl NBD C6-glucosylceramide (0.125 μM) into the LVI vials and mix with liver lipids.
- Load in carousel of HPLC system and analyze three times.
- Measure NBD glucosylceramide and ceramide of samples.
- Add 100 μl of methanol/chloroform into amber glass vials to reconstitute the extracted lipids of samples.
- Transfer 50 μl of reconstituted samples (~2,000 fluorescence units) into the LVI vials and load in carousel of HPLC system.
- Set up OpenLab CDS EZChrom program: flow rate 1 ml/min, injection volume 5 μl, linear gradient for 14 min (Solvent A 100%-0, and Solvent B 0-100%), and fluorescence detection (λ excitation 470 nm, λ emission 530 nm).
- Run HPLC analysis triplicate and save the files.
- Characterize and measure NBD C6-glucosylceramide with NBD C6-ceramide in standard solutions.
Data analysis
- Generate standard curves of Cer and GlcCer.
- Determine the average retention times for NBD C6-ceramide (Cer) and NBD C6-glucosylceramide (GlcCer) in chromatograms of standards opening in OpenLab CDS EZChrom program. Locate Cer at 3.10 ± 0.07 min and GlcCer at 3.8 ± 0.06 min in this column and solvent system.
- Transfer HPLC data (rslt.) from CDS EZChrom to Prism software. Define the AUC values at Y-axis and the corresponding pmols (per 5 μl, injected values) NBD C6-glucosylceramide (GlcCer) and C6-ceramide (Cer) at X-axis (Figure 1A).
- Generate the standard curves with equation (Y = MX + C) for GlcCer. Here, Y represents AUC (area under the curve, fluorescence units), X lipid amounts, M slope and C equation constant. For GlcCer, Y = 177.42X + 5.0143.
- Compare chromatograms of liver to confirm GlcCer in liver samples (Figure 3B).
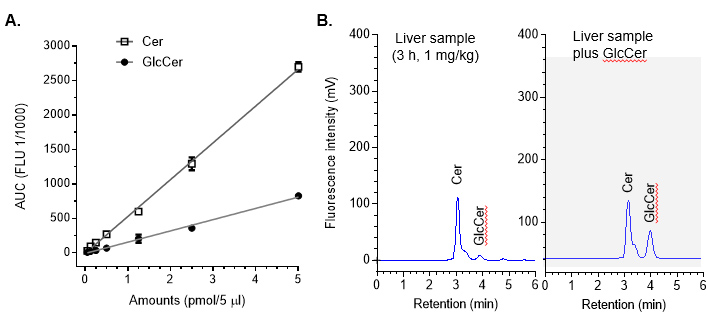
Figure 3. HPLC chromatogram of sphingolipids. A. Quantitation of C6-Cer and C6-GlcCer. Mixed standards of NBD C6-Cer and C6-GlcCer (1:1) was separated and quantitated by HPLC with fluorescence detection. The retention times for C6-Cer and C6-GlcCer were 3.1 and 3.8, respectively. Each data point represents mean ± SD of three independent experiments. B. Characterization of NBD C6-GlcCer in mouse liver. Lipids extracted from livers of mice after administration of NBD C6-Cer-RUB (1 mg/kg, i.p.; 180 min) were spiked with NBD C6-GlcCer (0.25 pmol per 5 μl) and analyzed by HPLC, respectively.
- Define the in vivo GCS activities of samples.
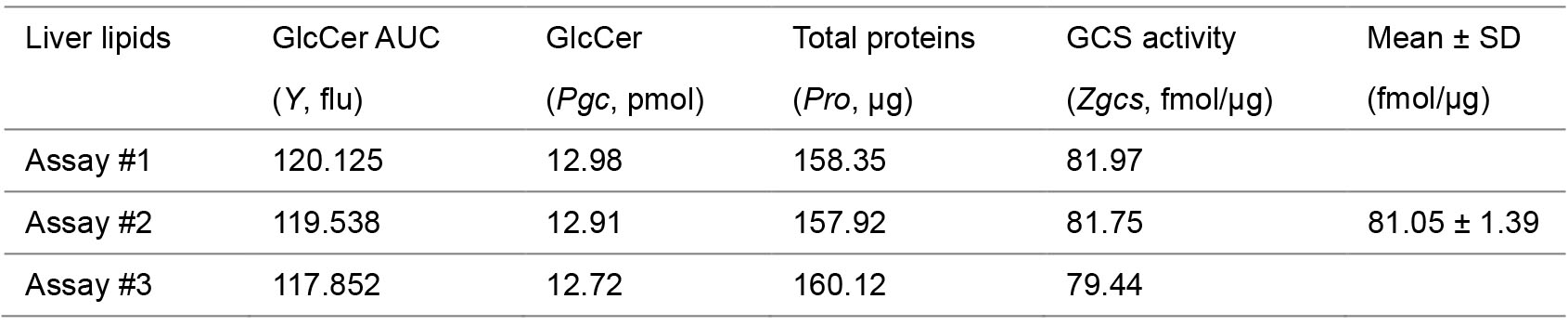
- Characterize Cer and GlcCer in the chromatographs of samples and calculate GlcCer amounts, with standard curves and equation. GlcCer amount (pmol/100 μl extracts), Pgc = 20 × X = 20 × (Y-5.0143)/177.42, (Figure 4A).
- Determine in vivo activities of GCS. The enzyme activity (fmol/μg) is briefly defined as the generated GlcCer (fmol) in 3 h by GCS in one μg of tissue proteins. Tissue GCS enzyme activities can be Zgcs = Pgc/Pro (Pgc from Data analysis step 2a, Pro from Step B4f) (Figure 4B).
- Example of calculation:
The in-vivo activities of GCS in tumors are significant higher than those in normal tissues, for instances of lung and liver (Figures 4B and 4C).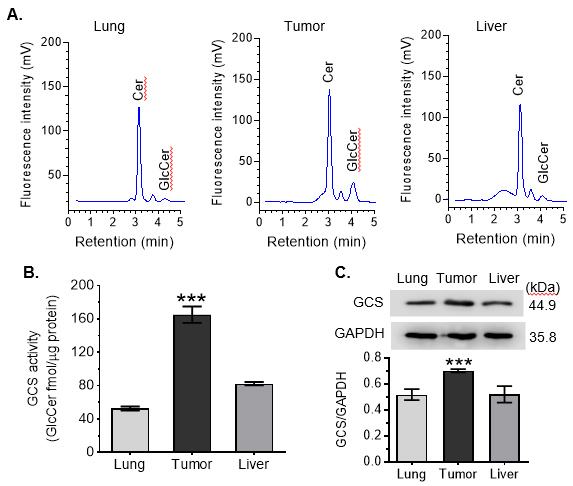
Figure 4. Intra-organ Cer glycosylation by GCS in mice. A. HPLC chromatograms. NBD Cer-RUB nanomicelles (1 mg/kg, i.p.; 4 cases/group) were administered to mice bearing an OVCAR-3 tumor xenograft; 3 h after injection, tissues were rapidly resected for lipid extraction and HPLC analysis. Approximately equal-fluorescence-unit samples of extracted sphingolipids were analyzed by HPLC. B. Intra-organ GCS activities. ***, P < 0.001 compared to lung or liver. C. Western blotting of tissue GCS. Equal amounts of proteins (50 μg protein/lane) extracted were resolved and then immunoblotted with antibodies for GCS or GAPDH. GCS protein levels are represented as ratios of GCS/GAPDH optical densities averaged from four blots. *, P < 0.05 compared to lung or liver.
Notes
- We take 80% of total lipid as well as total protein extracts in analyses and calculations of enzyme activities, which are 800 μl from 1 ml organic layer containing lipids, and 160 μl from 200 μl protein lysis buffer solution containing proteins, respectively.
- Handling NBD C6-ceramide or glucosylceramide as well as dissected tissues with light-proof, including using amber glass vials, aluminum covers and dim lights.
- NBD C6-ceramide complexed to BSA (Thermo Fisher Scientific, catalog number: N22651) can be used to analyze GCS activity in cells, as described previously (Gupta et al., 2010; Khiste et al., 2017).
- Intraparietal injection of NBD Cer-RUB (1 mg/kg) can provide sufficient substrates for ceramide glycosylation to measure in vivo GCS activities for most tissues, including tumor xenografts, except brain of mice.
- The analysis of NBD C6-Cer and NBD C6-GcLcer can also be carried on similar HPLC system with fluorescence detector from Waters, Shimadzu or other vendors. The retention time and detection sensitivity for NBD C6-GlcCer or NBD C6-Cer may vary with the solvents, column and HPLC system, but are still identical.
Recipes
- NBD Cer-RUB solution, 0.25 mg/ml
Add 25.28 mg NBD Cer-RUB nanomicelles (containing 0.5 mg NBD C6-ceramide) and 2 ml RPMI-1640 medium (serum-free) in a sterile 4-ml glass vial, vortex for 30 s, and keep at room temperature before animal injection - NBD Cer-RUB nanomicelles
NBD C6-ceramide (NBD Cer) is complexed with rubusoside (RUB) (1:50, w/w) to generate NBD Cer-RUB nanomicelles, as described previously (Khiste et al., 2017).
a.Dissolve NBD Cer in an ethanol solution containing RUB (140 g/100 ml)
b.Filter this solution through a 0.45-μm nylon filter
c.Evaporate and lyophilize to generate an NBD C6-Cer-RUB complex in powder form
d.Store NBD Cer-RUB powder at 4 °C - NP40 protein lysis solution
Add 198 μl NP40 Cell Lysis Buffer and 2 μl ProteaseArrestTM solution in 1.5 ml microcentrifuge tube, mix and keep it on ice before use - Solvent system for HPLC analysis
Solvent A:
Mix 400 ml chloroform, 100 ml methanol and 0.5 ml ortho-phosphoric acid in a 500 ml glass bottle and close it tightly
Solvent B:
Mix 300 ml chloroform, 170 ml methanol, 30 ml HPLC water and 0.5 ml ortho-phosphoric acid in a 500 ml glass bottle and close it tightly - NBD C6-ceramide or NBD C6-glucosylceramide standard solutions
10 μM NBD C6-ceramide (stock solution, -20 °C):
Add 10 μg NBD C6-ceramide (mw 575.75) and 1.737 ml chloroform/methanol (5:1, v:v) in a 4-ml amber glass vial, vortex and close it tightly
10 μM NDB C6-glucosylceramide (stock solution, -20 °C):
Add 10 μg N-hexanoyl-NBD-glucosylceramide (mw 738) and 1.355 ml chloroform/methanol (5:1, v:v) in a 4-ml amber glass vial, vortex and close it tightly
Note: Each stock solution will be diluted two or three times with chloroform/methanol and the diluted ones will be used to generate standard curves.
Acknowledgments
This work was supported by Public Health Service/NIH grant R15CA167476 from the National Cancer Institute (Y.Y.L) and P20 GM103424-11 from the National Institute of General Medical Sciences (Y.Y.L). This work was also partially supported by the Mizutani Foundation for Glycoscience, Japan (MFRG120068 Y.Y.L).
Competing interests
The authors declare no competing financial interests.
Ethics
All animal experiments were approved by the Animal Care and Use Committee of the University of Louisiana at Monroe (ULM) (Protocol# 16DEC-YYL 01) and were conducted in strict accordance with good animal practice as defined by NIH guidelines.
References
- Aerts, J. M., Ottenhoff, R., Powlson, A. S., Grefhorst, A., van Eijk, M., Dubbelhuis, P. F., Aten, J., Kuipers, F., Serlie, M. J., Wennekes, T., Sethi, J. K., O'Rahilly, S. and Overkleeft, H. S. (2007). Pharmacological inhibition of glucosylceramide synthase enhances insulin sensitivity. Diabetes 56(5): 1341-1349.
- Basu, M., De, T., Das, K. K., Kyle, J. W., Chon, H. C., Schaeper, R. J. and Basu, S. (1987). Glycolipids. Methods Enzymol 138: 575-607.
- Basu, S., Kaufman, B. and Roseman, S. (1968). Enzymatic synthesis of ceramide-glucose and ceramide-lactose by glycosyltransferases from embryonic chicken brain. J Biol Chem 243(21): 5802-5804.
- Bourteele, S., Hausser, A., Doppler, H., Horn-Muller, J., Ropke, C., Schwarzmann, G., Pfizenmaier, K. and Muller, G. (1998). Tumor necrosis factor induces ceramide oscillations and negatively controls sphingolipid synthases by caspases in apoptotic Kym-1 cells. J Biol Chem 273(47): 31245-31251.
- Chiu, W. H., Su, W. C., Li, C. L., Chen, C. L. and Lin, C. F. (2015). An increase in glucosylceramide synthase induces Bcl-xL-mediated cell survival in vinorelbine-resistant lung adenocarcinoma cells. Oncotarget 6(24): 20513-20524.
- D'Angelo, G., Capasso, S., Sticco, L. and Russo, D. (2013). Glycosphingolipids: synthesis and functions. FEBS J 280(24): 6338-6353.
- Fishman, P. H. and Brady, R. O. (1976). Biosynthesis and function of gangliosides. Science 194(4268): 906-915.
- Gupta, V., Bhinge, K. N., Hosain, S. B., Xiong, K., Gu, X., Shi, R., Ho, M. Y., Khoo, K. H., Li, S. C., Li, Y. T., Ambudkar, S. V., Jazwinski, S. M. and Liu, Y. Y. (2012). Ceramide glycosylation by glucosylceramide synthase selectively maintains the properties of breast cancer stem cells. J Biol Chem 287(44): 37195-37205.
- Gupta, V., Patwardhan, G. A., Zhang, Q. J., Cabot, M. C., Jazwinski, S. M. and Liu, Y. Y. (2010). Direct quantitative determination of ceramide glycosylation in vivo: a new approach to evaluate cellular enzyme activity of glucosylceramide synthase. J Lipid Res 51(4): 866-874.
- Hakomori, S. (1981). Glycosphingolipids in cellular interaction, differentiation, and oncogenesis. Annu Rev Biochem 50: 733-764.
- Hakomori, S. (2002). The glycosynapse. Proc Natl Acad Sci U S A 99(1): 225-232.
- Hayashi, Y., Horibata, Y., Sakaguchi, K., Okino, N. and Ito, M. (2005). A sensitive and reproducible assay to measure the activity of glucosylceramide synthase and lactosylceramide synthase using HPLC and fluorescent substrates. Anal Biochem 345(2): 181-186.
- Hosain, S. B., Khiste, S. K., Uddin, M. B., Vorubindi, V., Ingram, C., Zhang, S., Hill, R. A., Gu, X. and Liu, Y. Y. (2016). Inhibition of glucosylceramide synthase eliminates the oncogenic function of p53 R273H mutant in the epithelial-mesenchymal transition and induced pluripotency of colon cancer cells. Oncotarget 7(37): 60575-60592.
- Ichikawa, S., Sakiyama, H., Suzuki, G., Hidari, K. I. and Hirabayashi, Y. (1996). Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc Natl Acad Sci U S A 93(10): 4638-4643.
- Kasahara, K. and Sanai, Y. (2000). Functional roles of glycosphingolipids in signal transduction via lipid rafts. Glycoconj J 17(3 -4): 153-162.
- Khiste, S. K., Hosain, S. B., Dong, Y., Uddin, M. B., Roy, K. R., Hill, R. A., Liu, Z. and Liu, Y. Y. (2017). Incorporation of fluorescence ceramide-based HPLC assay for rapidly and efficiently assessing glucosylceramide synthase in vivo. Sci Rep 7(1): 2976.
- Lavie, Y., Cao, H., Volner, A., Lucci, A., Han, T. Y., Geffen, V., Giuliano, A. E. and Cabot, M. C. (1997). Agents that reverse multidrug resistance, tamoxifen, verapamil, and cyclosporin A, block glycosphingolipid metabolism by inhibiting ceramide glycosylation in human cancer cells. J Biol Chem 272(3): 1682-1687.
- Liu, Y. Y., Gupta, V., Patwardhan, G. A., Bhinge, K., Zhao, Y., Bao, J., Mehendale, H., Cabot, M. C., Li, Y. T. and Jazwinski, S. M. (2010). Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and β-catenin signaling. Mol Cancer 9: 145.
- Liu, Y. Y., Han, T. Y., Giuliano, A. E. and Cabot, M. C. (1999). Expression of glucosylceramide synthase, converting ceramide to glucosylceramide, confers adriamycin resistance in human breast cancer cells. J Biol Chem 274(2): 1140-1146.
- Liu, Y. Y., Han, T. Y., Giuliano, A. E. and Cabot, M. C. (2001). Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J 15(3): 719-730.
- Liu, Y. Y., Hill, R. A. and Li, Y. T. (2013). Ceramide glycosylation catalyzed by glucosylceramide synthase and cancer drug resistance. Adv Cancer Res 117: 59-89.
- Liu, Y. Y., Yu, J. Y., Yin, D., Patwardhan, G. A., Gupta, V., Hirabayashi, Y., Holleran, W. M., Giuliano, A. E., Jazwinski, S. M., Gouaze-Andersson, V., Consoli, D. P. and Cabot, M. C. (2008). A role for ceramide in driving cancer cell resistance to doxorubicin. FASEB J 22(7): 2541-2551.
- McEachern, K. A., Fung, J., Komarnitsky, S., Siegel, C. S., Chuang, W. L., Hutto, E., Shayman, J. A., Grabowski, G. A., Aerts, J. M., Cheng, S. H., Copeland, D. P. and Marshall, J. (2007). A specific and potent inhibitor of glucosylceramide synthase for substrate inhibition therapy of Gaucher disease. Mol Genet Metab 91(3): 259-267.
- Merrill, A. H., Jr. (2011). Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev 111(10): 6387-6422.
- Roh, J. L., Kim, E. H., Park, J. Y. and Kim, J. W. (2015). Inhibition of glucosylceramide synthase sensitizes head and neck cancer to cisplatin. Mol Cancer Ther 14(8): 1907-1915.
- Shukla, G. S. and Radin, N. S. (1990). Glucosyceramide synthase of mouse kidney: further characterization with an improved assay method. Arch Biochem Biophys 283(2): 372-378.
- Yoshizaki, F., Nakayama, H., Iwahara, C., Takamori, K., Ogawa, H. and Iwabuchi, K. (2008). Role of glycosphingolipid-enriched microdomains in innate immunity: microdomain-dependent phagocytic cell functions. Biochim Biophys Acta 1780(3): 383-392.
- Yu, R. K., Nakatani, Y. and Yanagisawa, M. (2009). The role of glycosphingolipid metabolism in the developing brain. J Lipid Res 50 Suppl: S440-445.
- Zhang, F., Koh, G. Y., Hollingsworth, J., Russo, P. S., Stout, R. W. and Liu, Z. (2012). Reformulation of etoposide with solubility-enhancing rubusoside. Int J Pharm 434(1-2): 453-459.
- Zhao, H., Przybylska, M., Wu, I. H., Zhang, J., Siegel, C., Komarnitsky, S., Yew, N. S. and Cheng, S. H. (2007). Inhibiting glycosphingolipid synthesis improves glycemic control and insulin sensitivity in animal models of type 2 diabetes. Diabetes 56(5): 1210-1218.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Roy, K. R., Khiste, S. K., Liu, Z. and Liu, Y. (2019). Fluorescence HPLC Analysis of the in-vivo Activity of Glucosylceramide Synthase. Bio-protocol 9(12): e3269. DOI: 10.21769/BioProtoc.3269.
Category
Cancer Biology > Cancer biochemistry > Drug resistance
Cell Biology > Cell metabolism > Lipid
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











