- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Organelle-associated rRNA Degradation
Published: Vol 9, Iss 11, Jun 5, 2019 DOI: 10.21769/BioProtoc.3255 Views: 6226
Reviewed by: Vishal S ParekhAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Import Assay into Mitochondria Isolated from Human Cells
Lena M. Murschall [...] Jan Riemer
Jun 20, 2021 6324 Views

Endoplasmic Reticulum Isolation: An Optimized Approach into Cells and Mouse Liver Fractionation
Marc Leiro [...] María Isabel Hernández-Alvarez
Sep 5, 2023 3504 Views

Analysis and Quantification of the Mitochondrial–ER Lipidome
Alexis Diaz-Vegas [...] James G. Burchfield
Jul 5, 2024 2579 Views
Abstract
Cytosolic rRNAs are highly dynamic and can be degraded under conditions such as apoptosis, starvation and magnesium depletion. The degradation is also related to their specific localization, as fractions of cytosolic ribosomes are localized on the surfaces of intracellular organelles, such as endoplasmic reticulum (ER) and mitochondria. Such localized translation facilitates translocation of nascent proteins into these organelles co-translationally, contributing to fast responses to cellular stresses and precise regulations of the organelle. Here, we describe a protocol to establish the in organello system to investigate rRNA degradation on mitochondrial outer membrane or ER. The protocol consists of organelle isolation, rRNA degradation on organelles and agarose gel electrophoresis to examine the remaining rRNAs.
Keywords: In organelloBackground
Cytosolic ribosomes, where protein translation takes place, have been shown to be localized to specific membranes within the cells, such as ER (Reid and Nicchitta, 2012) and mitochondrial outer membrane (Kellems and Butow, 1972), which couples protein synthesis to protein targeting and translocation (Mukhopadhyay et al., 2004). Such localized translation reduces the protein transportation cost and avoids mistargeting (Lesnik et al., 2015). In addition, localized translation contributes to fast responses to unfolded protein stress on ER (Reid et al., 2014) and modulates protein translation within mitochondria (Dennerlein et al., 2017; Richter-Dennerlein et al., 2016). The binding between cytosolic ribosomes and mitochondrial outer membrane have been previously investigated, and GTP, specific targeting sequences (Crowley and Payne, 1998) and MDI (Zhang et al., 2016) are all shown to be involved. However, how the mitochondrial outer membrane-associated cytosolic rRNAs are regulated by different conditions or mitochondrial proteins remains unclear, and how the ER-associated ribosomes differ from other pools of cytosolic ribosomes is not well studied. To gain a better understanding of these processes and their regulations in mammals, in organello systems are needed. However, no such systems have been established. Here, we describe a protocol to study in organello rRNA degradation on mitochondria or ER. The sample preparation procedures are minimized to reduce the operational errors. However, since this is an in organello system, the rRNA degradation observed may not reflect the real complexity in the cell. Particular caution needs to be taken in interpreting the results.
Materials and Reagents
- 1.5 ml microcentrifuge tubes (Quality Scientific Plastics, catalog number: 509-GRD-Q)
- 0.22 μm filter (Merk, Millex-GP PES, catalog number: SLGP033RB)
- Nuclease-free pipette tips (Quality Scientific Plastics, catalog numbers: T104RLS-Q, T090RLS-Q, and T112NXLRLS-Q)
- Mannitol (AMRESCO, catalog number: 0122-500G)
- Sucrose (AMRESCO, catalog number: 0335-500G)
- HEPES free acid (AMRESCO, catalog number: 0511-1KG)
- Sodium dodecyl sulfate (SDS) (AMRESCO, catalog number: 0227-1KG)
- EDTA, disodium salt, dihydrate (Na2EDTA·2H2O) (AMRESCO, catalog number: 0105-1KG)
- Agarose (BIOWEST, Regular Agarose G-10, catalog number: 111860)
- Proteinase K (AMRESCO, catalog number: 0706-100MG)
- Bromophenol blue (AMRESCO, ACS grade, catalog number: 0449-25G)
- Tris (AMRESCO, catalog number: 0497-5KG)
- Glacial acetic acid (Beijing Chemical Works, Analytic Reagent grade)
- Nuclease-free water (AMRESCO, catalog number: E476-1L)
- Glycerol (AMRESCO, catalog number:0854-1L)
- KOH (Sigma-Aldrich, catalog number: P1767-500G)
- Double distilled water (ddH2O)
- NaOH (sodium hydroxide pellets) (Shanghai Sangon Biotech, catalog number: A100173)
- Tween20 (AMRESCO, product code: 0777-1L)
- NaCl (AMRESCO, catalog number: 0241-1KG)
- PBS (Corning, catalog number: 21-040-CVR)
- Luminol/Enhancer solution (Thermo Fisher Scientific, catalog number: 1863096)
- Anti-Mortalin antibody (Sigma-Aldrich, catalog number: G4045)
- Anti-Calnexin antibody (Cell Signaling Technology, catalog number: 2433S)
- Anti-Rabbit IgG (whole molecule)–Peroxidase (Sigma-Aldrich, catalog number: A0545)
- Prestained Protein Ladder (Thermo Fisher Scientific, PageRuler, catalog number: 26616)
- HEK293 (from Carla M. Koehler’s lab at University of California, Los Angeles)
- 10% SDS-PAGE gel (Homemade, refer to He, 2011 for detailed protocol)
- Skim milk powder (OXOID, catalog number: LP0031)
- BSA (Sigma-Aldrich, catalog number: P3761)
- MitoPrep buffer (see Recipes)
- 10% (w/v) SDS (see Recipes)
- 0.5 M EDTA (pH 8.0) (see Recipes)
- 2x DNA-SDS-EDTA buffer (see Recipes)
- Proteinase K (1 mg/ml) (see Recipes)
- 50x TAE (see Recipes)
- 1x TBS-T (see Recipes)
- 2x protein loading buffer (see Recipes)
Equipment
- Pipettes (RAININ, Pipet-Lite XLS)
- Two heating blocks (Hangzhou Allsheng Instruments, Product Name: dry bath incubator, catalog number: MK200-2)
- NanoDrop instrument (Thermo Fisher Scientific, NanoDrop 2000c Spectrophotometer)
- Power supply (Tanon, catalog number: EPS 300)
- Gel imaging system (Tanon, catalog number: 1600)
- Centrifuges (Thermo Fisher Scientific, models: Sorvall Legend Micro 21 and Micro 21R)
- pH meter (Sartorius, catalog number: PB-10)
- -80 °C freezer (Thermo Fisher Scientific)
- Nitrocellulose membrane (Merck, catalog number: HATF00010)
- Vertical electrophoresis bath (Tanon, catalog number: VE-180)
- Glass/Teflon homogenizer (Wheaton, catalog number: 358034)
- Incubator
- 4 °C refrigerator
- -20 °C freezer
Software
- Tanon imaging (Tanon MP) and processing (Tanon GIS) software
- ImageJ
- GraphPad Prism 5
- Microsoft PowerPoint 2010
Procedure
Note: Use nuclease-free tubes and tips (Materials and Reagents sections) for all the steps.
- Crude mitochondria and ER isolation
Note: The method for mitochondria isolation was adapted from a previously described protocol (Wang et al., 2015) with minor modifications. During organelle isolation, the pH value of MitoPrep buffer is 7.4 if not specified.- Collect HEK293 cells by centrifuging at 1,000 x g for 2 min at room temperature (RT). Discard the supernatant, and resuspend the cell pellet with 1 ml of PBS. Spin again at 1,000 x g for 2 min at RT. Discard the supernatant.
Note: HEK293 cells were cultured in a 15-cm cell culture dish. With a density of ~90%, ~3 x 107 cells were harvested. - Resuspend the cells in 1.5 ml ice-cold MitoPrep buffer and transfer the mixture to a 5 ml glass/Teflon homogenizer pre-cooled in an ice bath. Perform 30 strokes on ice.
- Transfer the homogenate into a 1.5 ml microcentrifuge tube and centrifuge at 800 x g for 5 min at 4 °C. Transfer the supernatant to a new microcentrifuge tube.
- Resuspend the pellet in 1.5 ml MitoPrep buffer. Perform 20 strokes on ice. Transfer the homogenate into a 1.5 ml microcentrifuge tube and centrifuge at 800 x g for 5 min at 4 °C. Transfer the supernatant to a new microcentrifuge tube.
- Spin the supernatants from Steps1c and 1d at 800 x g for 5 min at 4 °C.
- Transfer the supernatants to new microcentrifuge tubes and centrifuge at 11,000 x g for 5 min at 4 °C. The resulting pellets are crude mitochondria.
- The supernatants are transferred into two new microcentrifuge tubes and spun at 21,000 x g for 10 min at 4 °C. The pellets are crude ER.
- Combine the pellets of mitochondria or ER from the two tubes by resuspending them in 1 ml MitoPrep buffer. Spin at 11,000 x g for 5 min or 21,000 x g for 10 min to spin down mitochondria or ER respectively.
- Discard the supernatant and resuspend mitochondria or ER with 30 μl of MitoPrep buffer (pH 7.4).
Note: Optiprep gradients can be used for further purification, if purer fractions are needed. - Measure protein concentrations with a NanoDrop instrument after diluting 1 μl of protein sample with 19 μl of 0.6% (w/v) SDS. The original protein concentrations are the measured values multiplied by 20.
Note: After we add 1 μl of protein sample to 19 μl of 0.6% SDS, the total volume becomes 20 μl, that means the concentration of this diluted protein sample is 1/20 of its original concentration. So, we can calculate the original protein concentration by a factor of 20. Usually, 3 x 107 cells can yield 800 μg of mitochondria and 150 μg of ER.
- Collect HEK293 cells by centrifuging at 1,000 x g for 2 min at room temperature (RT). Discard the supernatant, and resuspend the cell pellet with 1 ml of PBS. Spin again at 1,000 x g for 2 min at RT. Discard the supernatant.
- Take out 200 μg of mitochondria (in about 15 μl of MitoPrep buffer, pH 7.4), or 50 μg of ER (in about 10 μl of MitoPrep buffer, pH 7.4), and bring the volume to 60 μl with MitoPrep buffer with a pH value of interest (pH 7.4 and 6.5 are the two values we routinely use). Keep the samples on ice.
Notes:- Metal ions, metal chelators, or other conditions can be added to the sample before the next step. Some conditions that have effects on the degradation of mitochondrion-associated cytosolic rRNAs have been listed in Table 1.
Table 1. Conditions that affect the degradation of mitochondrion-associated cytosolic rRNAs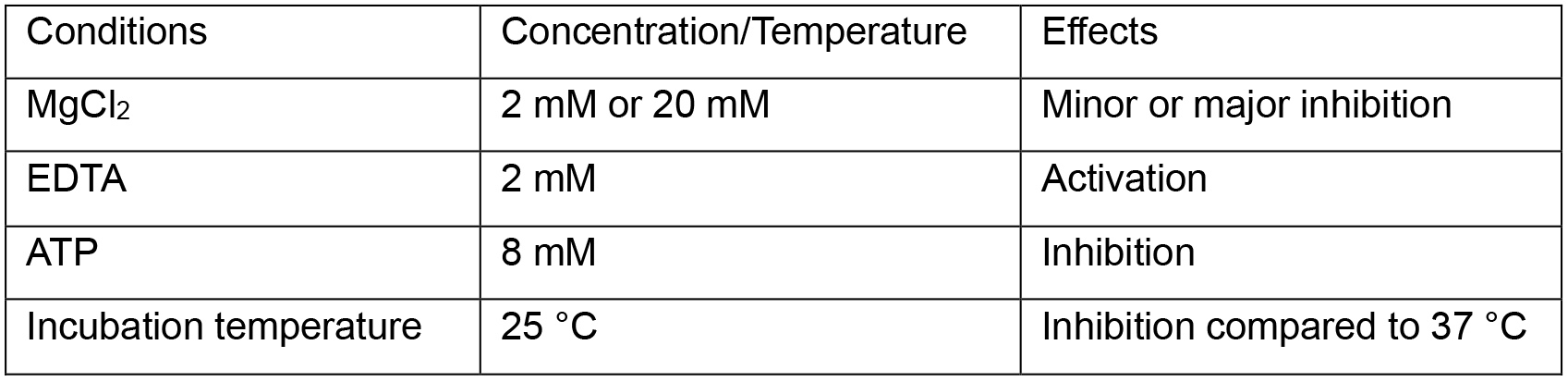
- For ER-associated rRNA degradation, the pH value of MitoPrep buffer matters: when the pH is 7.4, degradation is slower; when the pH is lowered to 6.5, the degradation becomes much faster. All these effects have been described in the original paper (Huang et al., 2018).
- Metal ions, metal chelators, or other conditions can be added to the sample before the next step. Some conditions that have effects on the degradation of mitochondrion-associated cytosolic rRNAs have been listed in Table 1.
- Divide each sample (60 μl) into 3 microcentrifuge tubes (20 μl per tube). Store one tube at the -80 °C freezer as the first time point (0 min). Transfer the other 2 tubes to 37 °C in a heating block. Incubate the two samples for 30 and 60 min respectively. At each time point, transfer one tube to -80 °C freezer. Store the samples at -80 °C for at least 15 min before the next step.
Note: For quality control, prepare another set of the samples and perform the same degradation steps. Then mix the samples with an equal volume of 2x protein loading buffer with DTT, and heat them at 95 °C for 5 min. Separate the proteins in the samples on a 10% SDS-PAGE gel. After running the gel at 150 V for 60 min, transfer the proteins in the gel to a nitrocellulose membrane at 450 mA for 70 min and blot the membrane by standard western blotting method. In brief, incubate the membrane at RT for 1 h in 5% skim milk, probe it with primary antibody in 1x TBS-T at RT for 1 h, wash 4 times with 1x TBS-T (5 min/time), incubate with secondary antibody in 1x TBS-T at RT for 1 h, and then wash 4 times with 1x TBS-T (5 min/time). Use Luminol/Enhancer solution for detection of the proteins. Marker proteins used for mitochondria and ER fractions are Mortalin (73.7 kDa) and Calnexin (~68 kDa) respectively. The actual position for Mortalin on a gel is between 70 and 100 kDa, and the position for Calnexin is around 100 kDa. - Take out samples from the -80 °C freezer and add 20 μl of 2x DNA-SDS-EDTA buffer. Vortex, and incubate the samples at 70 °C for 5 min. Cool the samples to RT.
- Add 0.5 μl of 1 mg/ml proteinase K to each sample and incubate it at 37 °C in a heating block for 5 min.
- Run the samples on a 1.5% (w/v) agarose gel with ethidium bromide (EB) (0.5 μg/ml) for 15 min at 150 V in 1x TAE buffer. Capture gel images to check RNA degradation pattern with a UV-based gel imaging system.
- The flow sheet of this protocol is shown in Figure 1.
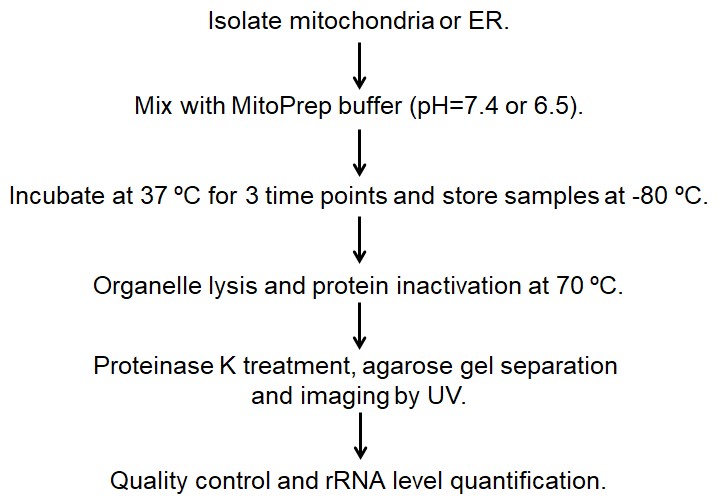
Figure 1. The flow sheet of this protocol
Data analysis
Representative data (Figures 2-4)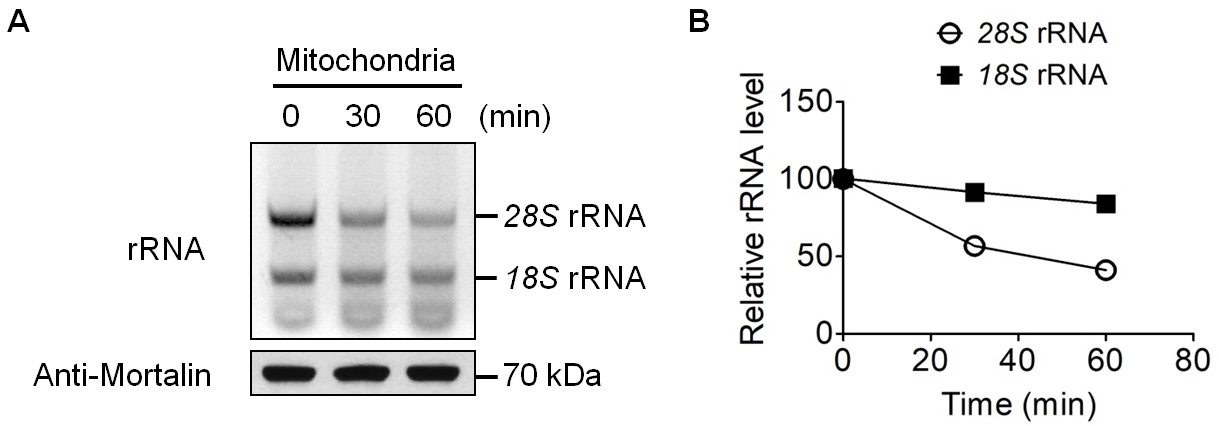
Figure 2. In organello degradation of mitochondrion-associated cytosolic rRNAs. A. Mitochondria were suspended in MitoPrep buffer and incubated for 0, 30 or 60 min. The top panel shows the agarose gel image of mitochondrion-associated cytosolic rRNAs at three time points. The two major bands are cytosolic 28S rRNA and 18S rRNA. The bottom panel is an immunoblot of mitochondrial matrix protein Mortalin (Mw: 73.7 kDa), which is used as a loading control. B. Quantification of the rRNAs in Panel A.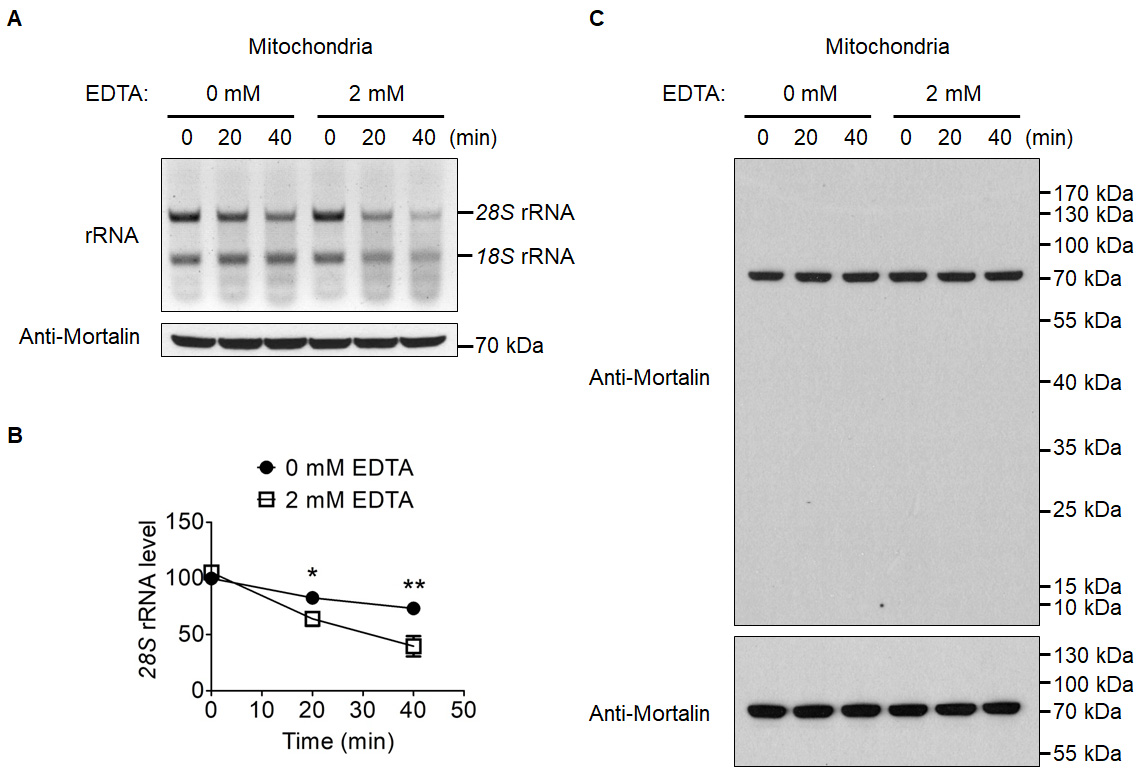
Figure 3. The effects of EDTA on the in organello degradation of mitochondrion-associated cytosolic rRNAs. A. Mitochondria were suspended in MitoPrep buffer, and incubated for 0, 20 or 40 min with or without 2 mM EDTA. The top panel shows the agarose gel image of mitochondrion-associated cytosolic rRNAs at three time points. The two major bands are cytosolic 28S rRNA and 18S rRNA. The bottom panel is an immunoblot of mitochondrial matrix protein Mortalin (Mw: 73.7 kDa), which is used as a loading control. B. Quantification of 28S rRNA in Panel A. C. Antibody specificity confirmation, protein loading control samples from A were separated with SDS-PAGE gels and transferred to nitrocellulose membrane. The entire membrane (upper panel) or the cropped membrane (bottom panel) was probed with anti-Mortalin antibody.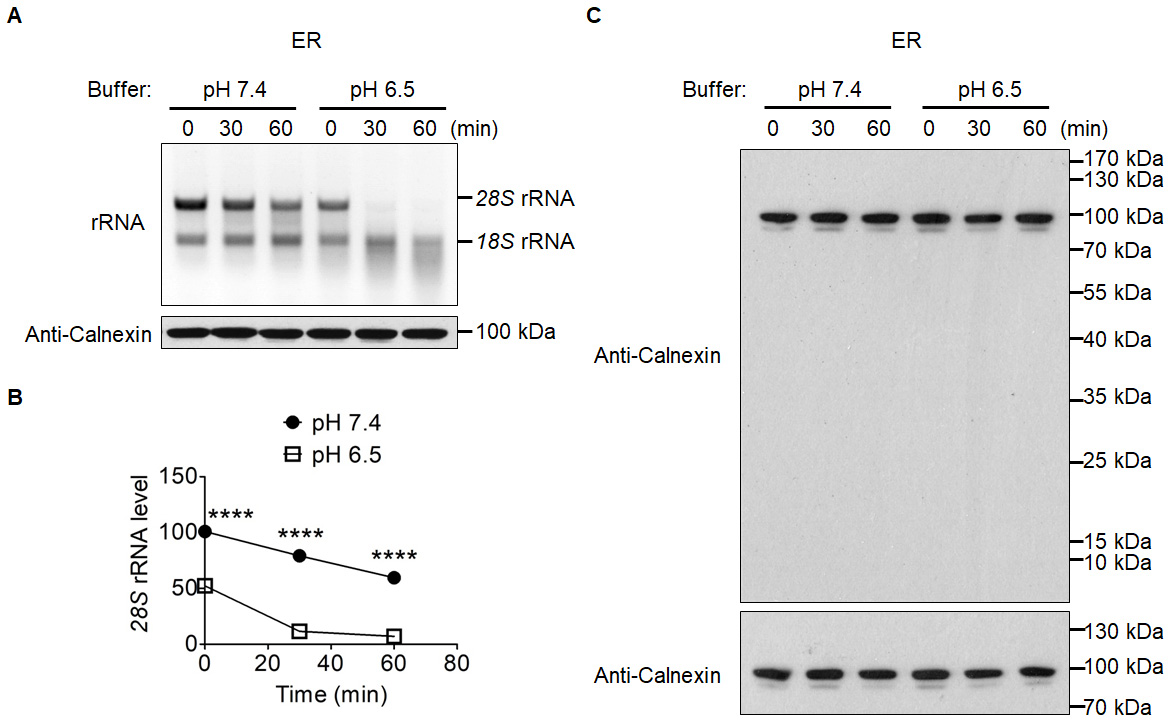
Figure 4. Effects of pH on the in organello degradation of ER-associated cytosolic rRNAs. A. ER was suspended in MitoPrep buffer with a pH of 7.4 or 6.5, and incubated for 0, 30 or 60 min. The top panel shows the agarose gel image of ER-associated cytosolic rRNAs at three time points. The two major bands are cytosolic 28S rRNA and 18S rRNA. The bottom panel is an immunoblot of ER protein Calnexin (the position of the protein on the gel is around 100 kDa), which is used as a loading control. B. Quantification of 28S rRNA in Panel A. C. Antibody specificity confirmation, protein loading control samples from A were separated with SDS-PAGE gels and transferred to nitrocellulose membrane. The entire membrane (upper panel) or the cropped membrane (bottom panel) was probed with Anti-Calnexin antibody.
Data processing
The gel image was captured with Tanon 1600 Gel Image System (Tanon), and cropped using Photoshop.
Data analysis
The intensity of the bands on each gel was measured with ImageJ, normalized to the protein loading control, and analysed with GraphPad Prism.
For an original image captured on Gel Image System (the background is black, while the bands are white), here is a brief introduction on how to use ImageJ for quantification. Open software ImageJ. Select “Rectangular” tool (this is default choice), and draw a rectangle covering the RNA band at the first time point. Press shortcut “Ctrl + M” to measure the band intensity. Move the same rectangle horizontally to RNA band at the second or third time point, and then press shortcut “Ctrl + M” to measure the band intensity. After all the time points are measured, move the same rectangle to a typical background area, and then press shortcut “Ctrl + M” to measure background intensity.
Calculation method: Measure the RNA signal by subtracting the background intensity from the band intensity to get the absolute intensity of each band. Set the signal at the first time point as 100, and calculate the second and third ones by dividing the absolute signal of the RNA band to that of the first time point.
Statistical comparisons were performed using unpaired t-tests (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are presented as mean ± standard deviation (S.D.). Data sets at each time point were analyzed individually.
Recipes
- MitoPrep buffer
0.225 M mannitol
0.075 M sucrose
20 mM HEPES (pH 7.4 or pH 6.5)- Dissolve 2.05 g of mannitol (Mw: 182.17), 1.28 g of sucrose (Mw: 342.3), and 0.2383 g of HEPES (Mw: 238.3) in ~45 ml ddH2O
- Adjust pH to 7.4 or 6.5 with 1 M KOH, and add ddH2O to the final volume of 50 ml
- Filter through 0.22 μm filter, and store at 4 °C
- 10% (w/v) SDS
- Dissolve 10 g of SDS in 100 ml of ddH2O, stir until completely dissolved. Store at RT
- Dilute 10% SDS to 0.6% SDS with ddH2O. Store at RT
- 0.5 M EDTA (pH 8.0)
- Add 9.306 g of EDTA (Mw: 372.24) to ~40 ml of ddH2O, stir and slowly adjust the pH to 8.0 with NaOH, and make up to 50 ml with ddH2O
- Filter through a 0.22 μm filter and store at 4 °C
- 2x DNA-SDS-EDTA buffer
2x DNA loading buffer, supplemented with 0.5% SDS and 15 mM EDTA
First prepare 10x DNA loading buffer:
For a 10 ml solution, dissolve 0.2 g of SDS and 0.01 g of bromophenol blue in 5 ml of ddH2O, mix with 5 ml of glycerol. Store at RT
To prepare 100 μl 2x DNA-SDS-EDTA buffer:
Mix 72 μl of nuclease-free water with 20 μl of 10x DNA loading buffer, add 5 μl of 10% SDS and 3 μl of 0.5 M EDTA (pH 8.0) - Proteinase K (1 mg/ml)
Dissolve 1 mg of proteinase K in 1 ml of nuclease-free water, aliquot and store at -20 °C - 50x TAE
2 M Tris
1 M acetic acid
50 mM EDTA- Dissolve 242 g of Tris (Mw: 121.14), 37.2 g of Na2EDTA·2H2O (Mw: 372.24) and 57.1 ml of glacial acetic acid (molar concentration: 17.5 M) in ~900 ml of ddH2O, add ddH2O to the final volume of 1 L. Store at RT
- Prepare 1x TAE by diluting 50x TAE with ddH2O
- 1x TBS-T
- Dilute 10x TBS stock with 0.1% (v/v) of Tween20 and ddH2O to make 1x TBS-T
- To prepare 10x TBS (1 L), dissolve 24.2 g of Tris and 80 g of NaCl in ~900 ml of ddH2O, and adjust the pH to 7.6 with HCl
- 2x protein loading buffer
- First prepare 100 ml of 0.5 M Tris-HCl (pH 6.8). Dissolve 6.05 g of Tris (Mw: 121.14) in ~90 ml of ddH2O, adjust pH to 6.8 with HCl. Add ddH2O to the final volume of 100 ml. Store at RT
- To prepare 10 ml 2x protein loading buffer, mix 1.25 ml of 0.5 M Tris-HCl (pH 6.8), 0.2 ml of 0.5% (w/v) bromophenol blue, 2.5 ml of glycerol, 2 ml of 10% SDS and 3.55 ml of ddH2O, store at RT
- A final concentration of 30 mM DTT is added in the 2x protein loading buffer right before use
Acknowledgments
This work was supported by Grant 2017YFA0504600 from the Priority Research Program of the Ministry of Science and Technology of China, by Grants 31371439 and 91649103 from the National Natural Science Foundation of the People’s Republic of China, and by funds from the Ministry of Education of the People’s Republic of China 1000 Talents Youth Program. This protocol was modified from Huang et al. (2018).
Competing interests
The authors declare no conflicts of interest with the contents of this article.
References
- Crowley, K. S. and Payne, R. M. (1998). Ribosome binding to mitochondria is regulated by GTP and the transit peptide. J Biol Chem 273(27): 17278-17285.
- Dennerlein, S., Wang, C. and Rehling, P. (2017). Plasticity of mitochondrial translation. Trends Cell Biol 27(10): 712-721.
- He, F. (2011). Laemmli-SDS-PAGE. Bio-protocol 1(11): e80.
- Huang, J., Liu, P. and Wang, G. (2018). Regulation of mitochondrion-associated cytosolic ribosomes by mammalian mitochondrial ribonuclease T2 (RNASET2). J Biol Chem 293(51): 19633-19644.
- Kellems, R. E. and Butow, R. A. (1972). Cytoplasmic-type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J Biol Chem 247(24): 8043-8050.
- Lesnik, C., Golani-Armon, A. and Arava, Y. (2015). Localized translation near the mitochondrial outer membrane: An update. RNA Biol 12(8): 801-809.
- Mukhopadhyay, A., Ni, L. and Weiner, H. (2004). A co-translational model to explain the in vivo import of proteins into HeLa cell mitochondria. Biochem J 382(Pt 1): 385-392.
- Reid, D. W., Chen, Q., Tay, A. S., Shenolikar, S. and Nicchitta, C. V. (2014). The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158(6): 1362-1374.
- Reid, D. W. and Nicchitta, C. V. (2012). Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem 287(8): 5518-5527.
- Richter-Dennerlein, R., Oeljeklaus, S., Lorenzi, I., Ronsor, C., Bareth, B., Schendzielorz, A. B., Wang, C., Warscheid, B., Rehling, P. and Dennerlein, S. (2016). Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell 167(2): 471-483 e410.
- Wang, G., Shimada, E., Nili, M., Koehler, C. M. and Teitell, M. A. (2015). Mitochondria-targeted RNA import. Methods Mol Biol 1264: 107-116.
- Zhang, Y., Chen, Y., Gucek, M. and Xu, H. (2016). The mitochondrial outer membrane protein MDI promotes local protein synthesis and mtDNA replication. EMBO J 35(10): 1045-1057.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Huang, J. and Wang, G. (2019). Organelle-associated rRNA Degradation. Bio-protocol 9(11): e3255. DOI: 10.21769/BioProtoc.3255.
- Huang, J., Liu, P. and Wang, G. (2018). Regulation of mitochondrion-associated cytosolic ribosomes by mammalian mitochondrial ribonuclease T2 (RNASET2). J Biol Chem 293(51): 19633-19644.
Category
Molecular Biology > RNA > RNA degradation
Cell Biology > Organelle isolation > Mitochondria
Cell Biology > Organelle isolation > Endoplasmic reticulum
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










