- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Simple Method to Determine Protein Redox State in Arabidopsis thaliana
Published: Vol 9, Iss 11, Jun 5, 2019 DOI: 10.21769/BioProtoc.3250 Views: 7477
Reviewed by: Jian ChenNoelia ForesiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Histone Deubiquitination Assay in Nicotiana benthamiana
Shujing Liu and Lars Hennig
Mar 5, 2018 6695 Views

Detection of Disulfides in Protein Extracts of Arabidopsis thaliana Using Monobromobimane (mBB)
Shin‐nosuke Hashida and Maki Kawai-Yamada
Mar 5, 2019 6424 Views

In vitro Auto- and Substrate-Ubiquitination Assays
Hye Lin Park [...] Gyeong Mee Yoon
Apr 5, 2022 3546 Views
Abstract
Thiol-based redox regulation is a posttranslational protein modification that plays a key role in many biological aspects. To understand its regulatory functions, we need a method to directly assess protein redox state in vivo. Here we present a simple procedure to determine protein redox state in a model plant Arabidopsis thaliana. Our method consists of three key steps: (i) redox fixation by rapidly freezing plant tissues in the liquid nitrogen, (ii) labeling of thiol groups with the maleimide reagent, and (iii) protein detection by Western blotting. The redox state of a specific or given protein can be discriminated by the mobility change on SDS-PAGE with high sensitivity. This method provides a novel strategy to dissect the working dynamics of the redox-regulatory system in plants.
Keywords: ProteinBackground
In living cells, proteins are subject to several posttranslational modifications. The dithiol/disulfide interconversion of a redox-active Cys pair, referred to as the redox regulation, is involved in a wide range of biological processes. In plant chloroplasts, this regulatory function seems to be strongly influenced by light conditions, because it is driven by a photosynthetically-generated reducing power (Buchanan, 1980). While the molecular machineries for supporting redox-regulatory system in chloroplasts, such as the thioredoxin (Trx), have been well identified so far (Buchanan et al., 2002; Lemaire et al., 2007; Jacquot, 2018), its working dynamics under environmental fluctuations remains poorly characterized. To address this issue, we need a methodology for analyzing the redox state of protein in vivo. To date, the spectrophotometric enzyme assay using a protein mixture extracted from intact chloroplasts or leaves has been often challenged on the assumption that the activation state of redox-regulated proteins is linked to their reduction level (Scheibe et al., 1989; Yoshida et al., 2007; Thormählen et al., 2015; Vaseghi et al., 2018). However, it is difficult from this assay to accurately estimate the redox state of the corresponding protein. Furthermore, we cannot exclude the possibility of nonspecific redox change during sample preparations and/or enzyme assays. To overcome these problems, an alternative approach to discriminate the redox state of proteins in a direct way is highly desirable. Here, we provide a simple protocol for determining protein redox state in a model plant Arabidopsis thaliana. This method has allowed us to uncover dynamic redox behaviors of several chloroplast proteins under varying light conditions (Yoshida et al., 2014, 2015 and 2018; Yoshida and Hisabori, 2016a and 2018), underpinning high sensitivity and accuracy of this strategy. This method is also applicable to proteins located in other cellular compartments (Yoshida and Hisabori, 2016b).
Materials and Reagents
- 1.5 ml microcentrifuge tube (WATSON, catalog number: 131-7155C)
- 10 μl pipette tip (WATSON, catalog number: 110-201C)
- 200 μl pipette tip (WATSON, catalog number: 110-705C)
- 1,000 μl pipette tip (WATSON, catalog number: 110-804C)
- 96-well EIA/RIA plate (Corning, catalog number: 3590)
- Absorbent paper (ATTO, catalog number: CB-09A)
- Immun-Blot PVDF membrane (Bio-Rad, catalog number: 1620177)
- Arabidopsis thaliana plants (Ecotype: Columbia-0)
- Liquid nitrogen
- Distilled water
- Tris(hydroxymethyl)aminomethane (Tris) (Nacalai Tesque, catalog number: 35434-21)
- Sodium lauryl sulfate (SDS) (Nacalai Tesque, catalog number: 31607-65)
- Glycerol (Nacalai Tesque, catalog number: 17018-83)
- Protease inhibitor cocktail tablet “cOmplete” (Roche, catalog number: 11836145001)
- Bromophenol Blue (BPB) (Wako, catalog number: 029-02912)
- 4-acetamido-4’-maleimidylstilbene-2,2’-disulfonate (AMS) (Invitrogen, catalog number: A485)
- Glycine (Nacalai Tesque, catalog number: 17141-95)
- Sodium chloride (NaCl) (Nacalai Tesque, catalog number: 31320-05)
- Polyoxyethylene (20) sorbitan monolaurate (Wako, catalog number: 167-11515)
- Methanol (Nacalai Tesque, catalog number: 21914-74)
- Pierce BCA protein assay kit (Thermo Scientific, catalog number: 23225)
- SDS-PAGE protein marker (e.g., Bio-Rad, catalog number: 161-0373)
- ECL Prime blocking agent (GE Healthcare, catalog number: RPN418V)
- ECL Prime Western blotting detection reagent (GE Healthcare, catalog number: RPN2232) or ECL Select Western blotting detection reagent (GE Healthcare, catalog number: RPN2235)
- Primary antibody for protein of interest
- Secondary antibody for protein of interest
- Immunoreaction enhancer (e.g., Can Get Signal, Toyobo, catalog number: NKB-101)
- Protein extraction/thiol labeling solution (see Recipes)
- Electrophoresis buffer for SDS-PAGE (see Recipes)
- Transfer buffer for Western blotting (see Recipes)
- TTBS (see Recipes)
Equipment
- Plant growth chamber (e.g., TOMY, model: CFH-415)
- 10 μl pipette (e.g., P10, Gilson, catalog number: F144802)
- 20 μl pipette (e.g., P20, Gilson, catalog number: F123600)
- 200 μl pipette (e.g., P200, Gilson, catalog number: F123601)
- 1,000 μl pipette (e.g., P1000, Gilson, catalog number: F123602)
- Mortar (AS ONE, catalog number: 5-4054-01)
- Pestle (AS ONE, catalog number: 5-4055-01)
- Refrigerated microcentrifuge (e.g., TOMY, model: MX-307)
- Heating blocks (e.g., TAITEC, model: CTU-Neo)
- Microplate reader iMark (Bio-Rad, catalog number: 1681130JA)
- Power supply (e.g., ATTO, model: AE-8135)
- Equipment for SDS-PAGE (e.g., NIHON EIDO, model: NA-1012)
- Equipment for Western blotting (e.g., BIO CRAFT, model: BE-320)
- Luminescence image analyzer (e.g., Fujifilm, model: LAS-4000 mini)
Software
- Software for image processing and analysis (e.g., ImageJ, http://imagej.nih.gov/ij//)
Procedure
An overview of the procedure is given in Figure 1.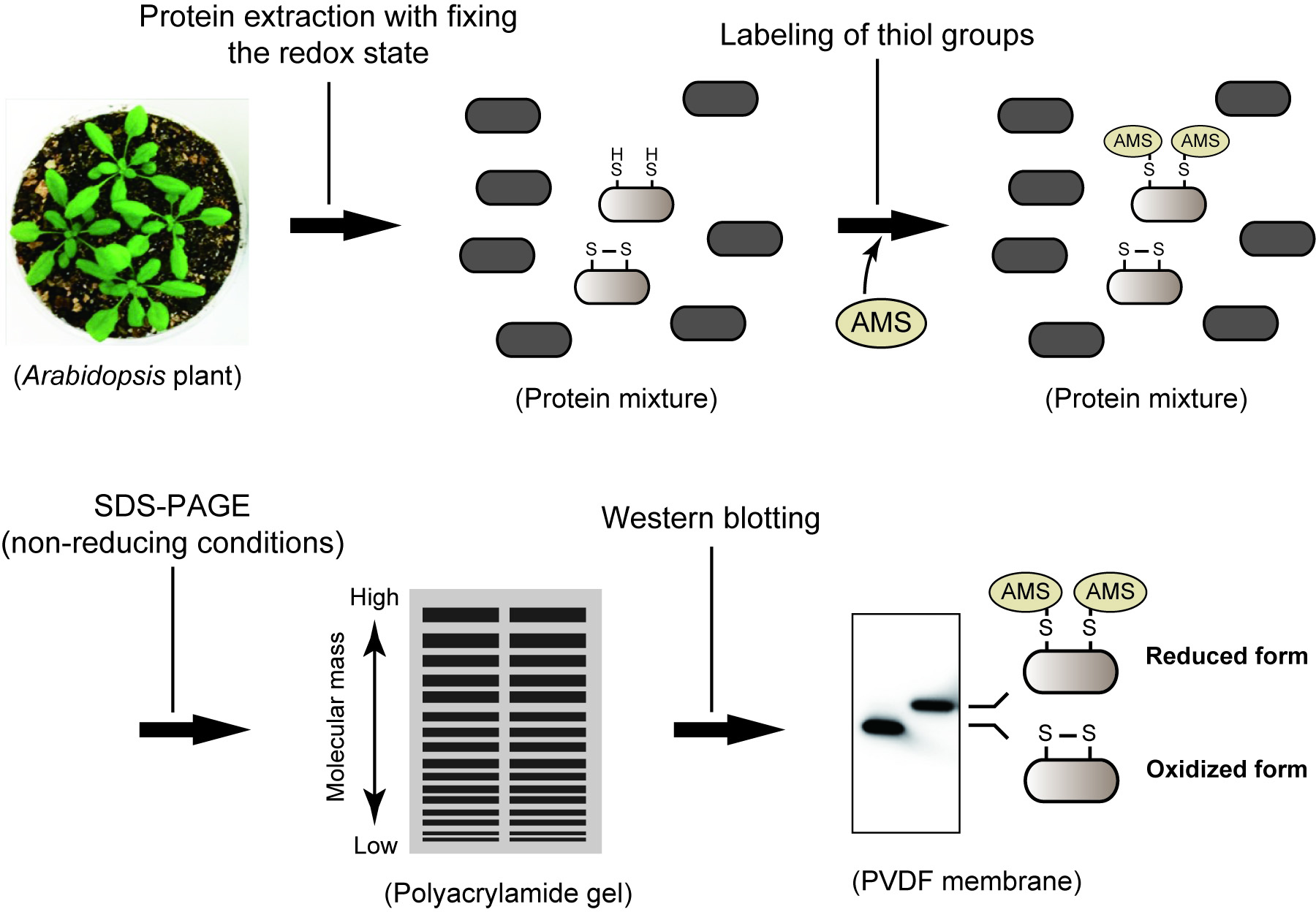
Figure 1. Simplified workflow for determining protein redox state in Arabidopsis thaliana
- Flash freezing of plant tissues, protein extraction, and thiol labeling
- Prepare the protein extraction/thiol labeling solution (see Recipes). Keep in the dark at room temperature.
- Place Arabidopsis plants under intended conditions (e.g., high light conditions) for intended periods (e.g., 10 min).
- Prepare the liquid nitrogen in a mortar just before the sampling.
- Excise plant tissues (e.g., leaves) and immediately immerse it in the liquid nitrogen in a mortar. This step should be done under intended conditions. We usually use two or three fully expanded leaves from plants grown under long-day (16-h day/8-h night) conditions for 4 weeks (50-100 mg of fresh weight).
- Grind plant tissues using a pestle. Transfer the powdered plant tissues to the microcentrifuge tube carefully.
- Add the protein extraction/thiol labeling solution. For our conditions described in Step A4, more than 300 μl of the solution (3-6 μl/mg leaf fresh weight) should be added. Then, keep in the dark at room temperature for 1 h to allow free thiol groups to be fully labeled with AMS. AMS is a maleimide reagent reactive with the free thiol, but not with the disulfide bond.
- Heat the protein sample (e.g., at 95 °C for 5 min).
- Centrifuge at 20,000 x g, room temperature for 10 min.
- Transfer the supernatant to a new microcentrifuge tube for removing the debris.
- Determine the protein concentration using the Pierce BCA protein assay kit (according to the manufacturer’s instructions). An equal amount of the protein extraction/thiol labeling solution should be added to the albumin standards. In our conditions described in Steps A4-A6, the protein concentration is estimated to be 4-6 mg/ml in most cases.
Note: Usually, the protein sample is stable at -30 °C for several months.
- SDS-PAGE and Western blotting
- Prepare the polyacrylamide gel for SDS-PAGE. We usually use the separating gel with a size of 85 x 60 x 1 mm.
Note: The optimal concentration of acrylamide is variable depending on the protein of interest. The molecular mass is a critical determinant. For example, we use the 10% acrylamide gel for detecting ATP synthase CF1-γ subunit (ca. 35,000) or fructose-1,6-bisphosphatase (FBPase; ca. 40,000), and the 15% acrylamide gel for detecting m-type Trx (ca. 12,000) (Figure 2). - Set the polyacrylamide gel to the electrophoresis tank. Pour the electrophoresis buffer (see Recipes).
- Load the protein sample (equivalent to 10-40 μg of proteins) to each well. Run the electrophoresis.
Note: The optimal amount for sample loading is variable depending on the protein of interest. The migration time for SDS-PAGE is important for better separation of proteins in the reduced and oxidized forms, which is also variable depending on the protein of interest. - Prepare the PVDF membrane and absorbent papers for Western blotting. The PVDF membrane must be hydrophilically treated with the methanol (for several seconds) and then the transfer buffer (for several minutes; see Recipes) prior to the use.
- Set the polyacrylamide gel after SDS-PAGE, the PVDF membrane, and the absorbent papers to the blotting equipment. Run the blotting.
- After the blotting, wash the PVDF membrane with distilled water and then TTBS (see Recipes) for several minutes.
- Incubate the PVDF membrane with 2% (w/v) blocking agent dissolved in TTBS (e.g., overnight at 4 °C).
- Incubate the PVDF membrane with the primary antibody diluted with TTBS (e.g., at room temperature for 2 h).
- Wash the PVDF membrane with TTBS (e.g., 10 min x 3 times).
- Incubate the PVDF membrane with the secondary antibody diluted with TTBS (e.g., at room temperature for 1 h).
- Wash the PVDF membrane with TTBS (e.g., 10 min x 3 times).
- Incubate the PVDF membrane with the ECL Prime solution. Detect the luminescence.
Note: The optimal conditions for the immunoreaction (Steps B6-B11) and the signal detection (Step B12) are largely variable depending on the protein of interest. The dilution ratio of antibodies should be carefully tested. If the signal is weak, it may be helpful to use the immunoreaction enhancer (e.g., Can Get Signal, Toyobo) or the high-sensitivity detection reagent (e.g., ECL Select, GE Healthcare).
- Prepare the polyacrylamide gel for SDS-PAGE. We usually use the separating gel with a size of 85 x 60 x 1 mm.
Data analysis
- The thiol-labeling maleimide reagent AMS has a molecular mass of 536.44 and, thereby, lowers protein mobility on SDS-PAGE. We can discriminate the redox state of objective protein based on an observable band shift (Figure 1). The scale of band shift is apparently variable depending on the protein of interest. If necessary, the reduction level of protein can be calculated by quantifying the signal intensity from the reduced and oxidized forms (using the image-analyzing software such as ImageJ).
- By applying this method to protein samples sequentially collected from changing environmental conditions (e.g., light), we can chase the redox behaviors of proteins under environmental fluctuations (Figure 2). Furthermore, a combined dataset for multiple proteins can provide insights into the working dynamics of the overall redox-based regulatory network in vivo (e.g., Yoshida et al., 2018).
- A constitutively expressed protein should be analyzed as the loading control. We usually analyze the Rubisco large subunit.
- Besides the formation of disulfide bond, some proteins undergo another type of oxidation, including sulfenylation and nitrosylation. To discriminate them, we need additional approaches such as the mass spectrometry-based analysis.
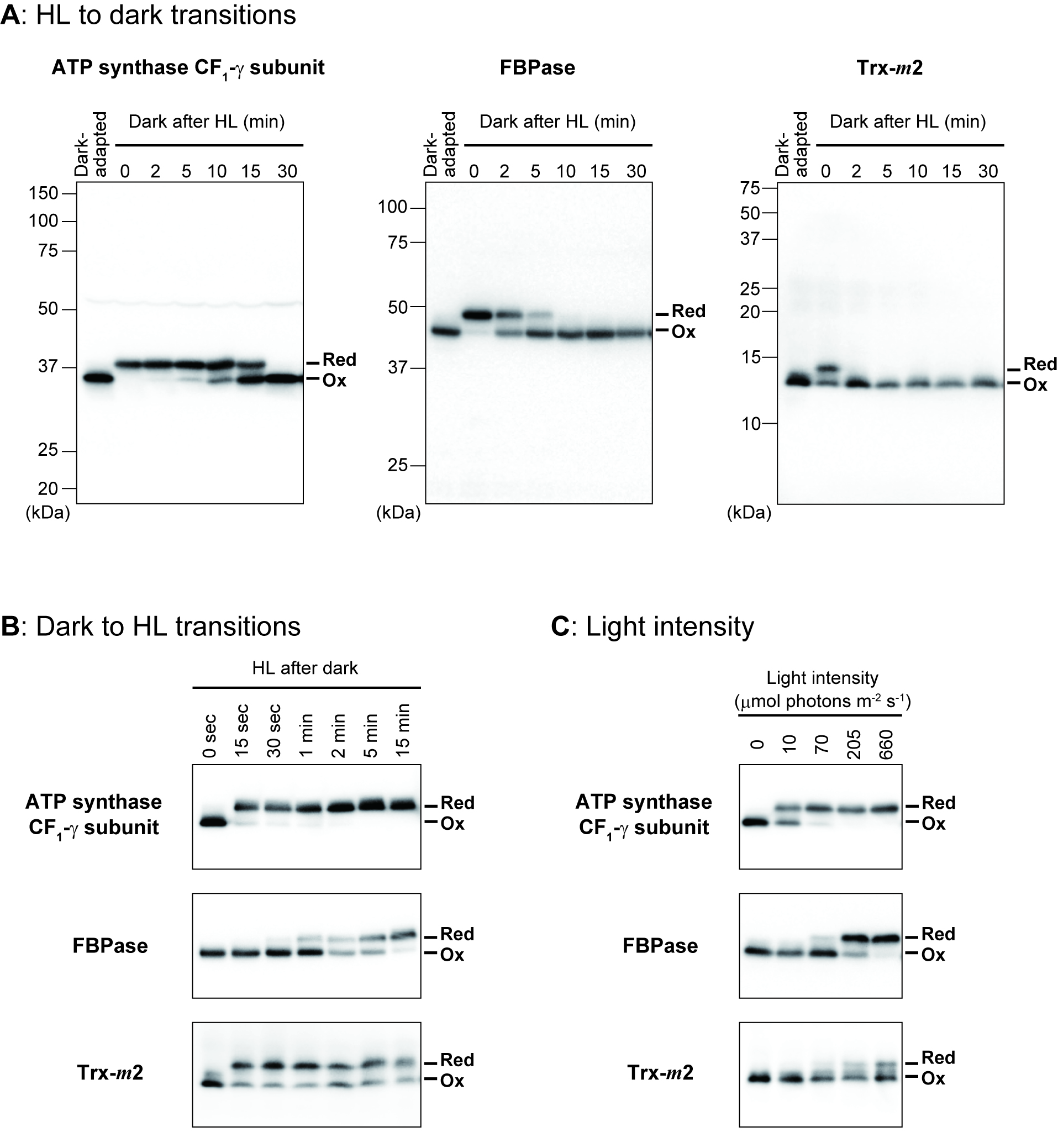
Figure 2. Examples for protein redox dynamics revealed by this protocol. Results of three chloroplast proteins (ATP synthase CF1-γ subunit, fructose-1,6-bisphosphatase (FBPase), and m-type Trx (Trx-m2)) are shown. The protein samples were prepared from Arabidopsis plants placed under high light (HL) to dark transitions (A), under dark to HL transitions (B), and at different light intensities for 15 min (C). Similar results are shown in our previous papers (A: Yoshida et al., 2018; B: Yoshida and Hisabori, 2018; C: Yoshida et al.; 2015). See each of these original papers for the detailed sampling conditions.
Recipes
- Protein extraction/thiol labeling solution
62.5 mM Tris-HCl (pH 6.8)
2% (w/v) SDS
7.5% (v/v) glycerol
0.01% (w/v) BPB
4 mM AMS
Note: AMS must be added just before the use. Protease inhibitor cocktail tablet “cOmplete” should be also added. Dissolve one tablet in 2 ml distilled water, which can be used as 25x stock solution. - Electrophoresis buffer for SDS-PAGE
25 mM Tris
0.1% (w/v) SDS
192 mM glycine - Transfer buffer for Western blotting
25 mM Tris
192 mM glycine
20% (v/v) methanol - TTBS
20 mM Tris-HCl (pH 7.5)
500 mM NaCl
0.1% (v/v) polyoxyethylene(20) sorbitan monolaurate
Acknowledgments
This protocol was adapted from Yoshida et al. (2018) and our other studies (see Background). This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant numbers: 26840090 to K.Y., 16H06556 to K.Y. and T.H.).
Competing interests
The authors declare that there is no conflict of interest.
References
- Buchanan, B. B. (1980). Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol 31: 341-374.
- Buchanan, B. B., Schürmann, P., Wolosiuk, R. A. and Jacquot, J. P. (2002). The ferredoxin/thioredoxin system: from discovery to molecular structures and beyond. Photosynth Res 73(1-3): 215-222.
- Jacquot, J. P. (2018). Dark deactivation of chloroplast enzymes finally comes to light. Proc Natl Acad Sci U S A 115(38): 9334-9335.
- Lemaire, S. D., Michelet, L., Zaffagnini, M., Massot, V. and Issakidis-Bourguet, E. (2007). Thioredoxins in chloroplasts. Curr Genet 51(6): 343-365.
- Scheibe, R., Geissler, A. and Fickenscher, K. (1989). Chloroplast glucose-6-phosphate dehydrogenase: Km shift upon light modulation and reduction. Arch Biochem Biophys 274(1): 290-297.
- Thormählen, I., Meitzel, T., Groysman, J., Öchsner, A. B., von Roepenack-Lahaye, E., Naranjo, B., Cejudo, F. J. and Geigenberger, P. (2015). Thioredoxin f1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant Physiol 169(3): 1766-1786.
- Vaseghi, M. J., Chibani, K., Telman, W., Liebthal, M. F., Gerken, M., Schnitzer, H., Mueller, S. M. and Dietz, K. J. (2018). The chloroplast 2-cysteine peroxiredoxin functions as thioredoxin oxidase in redox regulation of chloroplast metabolism. Elife 7: e38194.
- Yoshida, K., Hara, A., Sugiura, K., Fukaya, Y. and Hisabori, T. (2018). Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc Natl Acad Sci U S A 115(35): E8296-E8304.
- Yoshida, K., Hara, S. and Hisabori, T. (2015). Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J Biol Chem 290(23): 14278-14288.
- Yoshida, K. and Hisabori, T. (2016a). Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc Natl Acad Sci U S A 113(27): E3967-E3976.
- Yoshida, K. and Hisabori, T. (2016b). Adenine nucleotide-dependent and redox-independent control of mitochondrial malate dehydrogenase activity in Arabidopsis thaliana. Biochim Biophys Acta 1857(6): 810-818.
- Yoshida, K. and Hisabori, T. (2018). Determining the rate-limiting step for light-responsive redox regulation in chloroplasts. Antioxidants (Basel) 7(11): 153.
- Yoshida, K., Matsuoka, Y., Hara, S., Konno, H. and Hisabori, T. (2014). Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana. Plant Cell Physiol 55(8): 1415-1425.
- Yoshida, K., Terashima, I. and Noguchi, K. (2007). Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol 48(4): 606-614.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yoshida, K. and Hisabori, T. (2019). Simple Method to Determine Protein Redox State in Arabidopsis thaliana. Bio-protocol 9(11): e3250. DOI: 10.21769/BioProtoc.3250.
Category
Plant Science > Plant biochemistry > Protein > Modification
Biochemistry > Protein > Modification
Biochemistry > Other compound > Thiol
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









