- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Improved Mammalian Mitochondrial RNA Isolation
Published: Vol 9, Iss 11, Jun 5, 2019 DOI: 10.21769/BioProtoc.3247 Views: 8325
Reviewed by: Aswad KhadilkarAlvaro Eduardo GalvisAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Updated Pseudo-seq Protocol for Transcriptome-Wide Detection of Pseudouridines
Yi Pan [...] Paul L. Boutz
May 5, 2024 1618 Views

Analysis and Quantification of the Mitochondrial–ER Lipidome
Alexis Diaz-Vegas [...] James G. Burchfield
Jul 5, 2024 2579 Views

An Optimized RNA Extraction Method From Micro-quantities of Guinea Pig Cartilage and Synovium for Osteoarthritis Research
Nidhi Bhardwaj [...] Jyotdeep Kaur
Jun 20, 2025 1607 Views
Abstract
Mitochondria have two sets of RNAs. One is encoded in mitochondrial genome, and the other that consists of imported RNAs within mitochondria and cytosolic RNAs associated with mitochondrial outer membrane is encoded in the nucleus. These mitochondrial RNAs play important roles in mitochondrion biosynthesis and signaling in and out of mitochondria. Isolation and analysis of mitochondrial RNAs can provide useful information on understanding the mitochondrial regulation of cellular processes. However, several ribonuclease activities have been found in mitochondria, which will degrade mitochondrial RNAs during the isolation process if they are not properly inactivated. Here, we describe an improved method to inactivate the ribonuclease activities prior to RNA extraction, and thus provide a reliable protocol to isolate mammalian mitochondrial RNAs for quantitative RT-PCR and other assays.
Keywords: Mitochondrial RNABackground
Mitochondrial RNAs are more diverse than originally thought. Firstly, each strand of mitochondrial DNA is transcribed as a long polycistronic transcript and processed to mature mRNAs, rRNAs and tRNAs (Hällberg and Larsson, 2014). Secondly, some nucleus-encoded noncoding RNAs are imported into mitochondria (Entelis et al., 2002; Wang et al., 2010; Mercer et al., 2011; Zhang et al., 2014; Cheng et al., 2018). In addition, cytosolic ribosomes and some mRNAs are found to be associated with mitochondrial outer membrane (Kellems and Butow, 1972; Williams et al., 2014). Analysis of mitochondrion-related RNAs can provide important information on understanding the mitochondrial biosynthesis and mitochondrial regulation of cellular processes, such as protein translation (Huang et al., 2018) and aging (Zheng et al., 2019). However, isolation of mitochondrial RNAs can be problematic since there are several strong ribonucleases within mitochondria (Rorbach et al., 2011; Bruni et al., 2013; Levy et al., 2016; Liu et al., 2017). Some of these ribonucleases remain highly active even under harsh conditions, such as proteinase-K treatment (Liu et al., 2017). To avoid partial degradation of mitochondrial RNAs after lysis of mitochondria, a reliable method to inactivate these ribonucleases during RNA isolation is needed. Most current mitochondrial RNAs isolation protocols, however, do not mention the potential consequences of incomplete RNase inactivation. Chloroform (Geiger and Dalgaard, 2018) or commercial kits (Sripada et al., 2012) are routinely used, but neither could completely inactivate the mitochondrial ribonucleases. In one protocol (Mager-Heckel et al., 2007), sodium dodecyl sulfate (SDS) and high temperature (100 °C) are used, but these conditions have a negative effect on RNA stability (Sidova et al., 2015). Here, we describe an improved method to inactivate the ribonuclease activities prior to RNA extraction. In this method, SDS lysis coupled with a milder temperature (70 °C) incubation denatures most proteins without causing RNA instability and the subsequent proteinase K digestion degrade nearly all proteins, providing a reliable protocol to isolate mammalian mitochondrial RNA for quantitative RT-PCR and other assays.
Materials and Reagents
- 1.5 ml microcentrifuge tubes (Quality Scientific Plastics, catalog number: 509-GRD-Q)
- 0.22 μm filter (Merk, Millex-GP PES, catalog number: SLGP033RB)
- Nuclease-free pipette tips (Quality Scientific Plastics, catalog numbers: T104RLS-Q, T090RLS-Q and T112NXLRLS-Q)
- Tris (AMRESCO, catalog number: 0497-5KG)
- Sodium dodecyl sulfate (SDS) (AMRESCO, catalog number: 0227-1KG)
- EDTA, disodium salt, dihydrate (Na2EDTA·2H2O) (AMRESCO, catalog number: 0105-1KG)
- Proteinase K (AMRESCO, catalog number: 0706-100MG)
- 6x DNA Loading Dye (Thermo Fisher Scientific, catalog number: R0611)
- Agarose (BIOWEST, Regular Agarose G-10, catalog number: 111860)
- Nuclease-free water (AMRESCO, catalog number: E476-1L)
- Double distilled water (ddH2O)
- DNase I, RNase-free, supplied with 10x Reaction buffer with MgCl2 (Thermo Fisher Scientific, catalog number: EN0521)
- TRIzol (Life Technologies, catalog number: 15596018)
- 1-Bromo-3-chloropropane (Sigma-Aldrich, catalog number: B9673)
- Isopropanol (AMRESCO, catalog number: 0918-1L)
- Ethanol (Sigma-Aldrich, catalog number: 459836-2L)
- HCl (Beijing Chemical Works, Analytic Reagent grade)
- NaOH (sodium hydroxide pellets) (Shanghai Sangon Biotech, catalog number: A100173)
- GeneRuler 1 kb DNA Ladder (Thermo Fisher Scientific, catalog number: SM0311)
- HEPES free acid (AMRESCO, catalog number: 0511-1KG)
- Mannitol (AMRESCO, catalog number: 0122-500G)
- Sucrose (AMRESCO, catalog number: 0335-500G)
- MitoPrep buffer (see Recipes)
- 10% (w/v) SDS (see Recipes)
- 1 M Tris-HCl (pH 7.4) (see Recipes)
- 0.5 M EDTA (pH 8.0) (see Recipes)
- Proteinase K (1 mg/ml) (see Recipes)
- Lysis buffer (see Recipes)
- 1x DNase buffer (see Recipes)
- 75% ethanol (v/v) (see Recipes)
Equipment
- Pipettes (RAININ, Pipet-Lite XLS)
- Two heat blocks (Hangzhou Allsheng Instruments, Product Name: dry bath incubator, catalog number: MK200-2)
- NanoDrop instrument (Thermo Fisher Scientific, NanoDrop 2000c Spectrophotometer)
- Power supply (Tanon, catalog number: EPS 300)
- Gel imaging system (Tanon, catalog number: 1600)
- Centrifuges (Thermo Fisher Scientific, models: Sorvall Legend Micro 21 and Micro 21R)
- pH meter (Sartorius, catalog number: PB-10)
- -80 °C freezer (Thermo Fisher Scientific)
- Magnetic stir bars and magnetic stirrer
- Fume hood
- Laminar flow cabinet
- Incubator
- 4 °C refrigerator
- -20 °C freezer
Procedure
The flow sheet of this protocol is shown in Figure 1.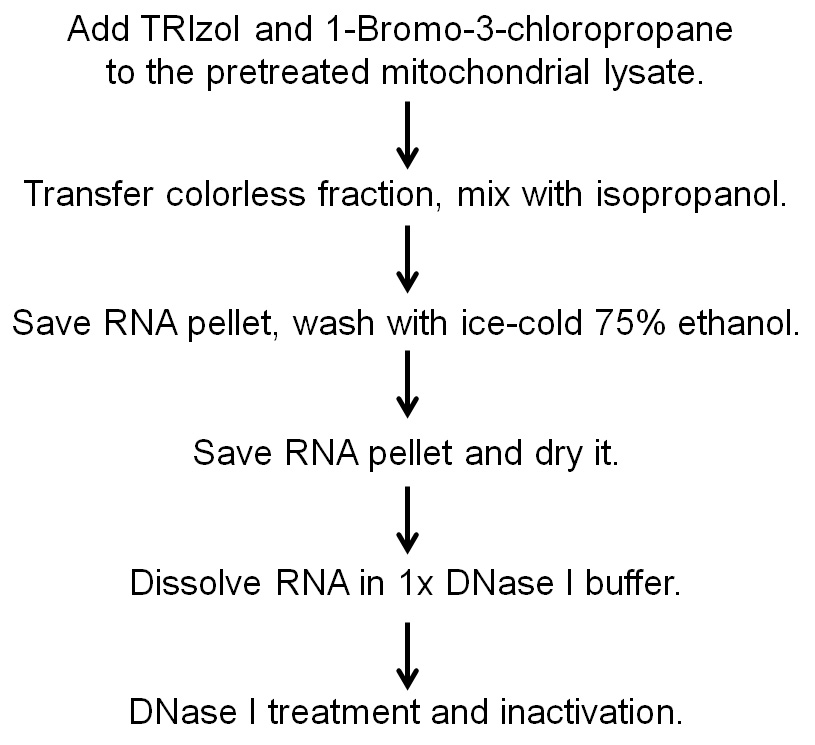
Figure 1. The flow sheet of steps in this protocol
- Sample pretreatment
Note: To examine sub-mitochondrially localized RNA, different treatments such as nuclease treatment or mitoplasting are needed. We focus here on inactivation of mitochondrial RNases. Other methods such as proximity labeling are also needed to separate mitochondrially localized RNA from RNA contamination from other cellular compartments.- Isolate mitochondria from tissue culture cells as described in Huang and Wang (2019) and measure mitochondrial protein concentration with a NanoDrop instrument after diluting 1 μl of mitochondrial sample with 19 μl of 0.6% SDS. Transfer 200 μg of purified mitochondria (in 20 μl of MitoPrep buffer) to a microcentrifuge tube and keep the tube on ice.
Note: About 9 x 106 HEK293 cells could be harvested from a 10-cm cell culture dish of 90% cell density, for a yield of around 250 μg mitochondria. - Mix mitochondria with 100 μl of fresh lysis buffer and heat the sample at 70 °C in a heat block for 5 min. Take out the sample, and let it cool down to room temperature (RT).
- Add 1 μl of 1 mg/ml proteinase K to the lysate and incubate it at 37 °C in a heat block for 5 min.
- Isolate mitochondria from tissue culture cells as described in Huang and Wang (2019) and measure mitochondrial protein concentration with a NanoDrop instrument after diluting 1 μl of mitochondrial sample with 19 μl of 0.6% SDS. Transfer 200 μg of purified mitochondria (in 20 μl of MitoPrep buffer) to a microcentrifuge tube and keep the tube on ice.
- RNA extraction
Note: Perform this part in a fume hood.- Add 400 μl of TRIzol and 200 μl of 1-Bromo-3-chloropropane to the pretreated lysate, shake the tube hard for 1 min, and then spin at 14,800 x g for 5 min at RT.
- Transfer the colorless fraction in the upper layer to a new microcentrifuge tube, mix with an equal volume of isopropanol and spin at 21,000 x g for 10 min at 4 °C.
- Discard the supernatant and save the RNA pellet. Add 600 μl of ice-cold 75% ethanol to the tube, gently turn it upside down for a couple of times, and spin at 21, 000 x g for 5 min at 4 °C.
Note: If a small amount of mitochondria (200 μg or less) was used, the RNA pellet might be very small and hardly visible. - Discard the supernatant and make the remaining ethanol as little as possible. Be careful to avoid losing the pellet.
- Dry the RNA pellet in the laminar flow cabinet.
- RNA treatment
Note: If mitochondrial DNA would not interfere with downstream assays, DNase I treatment is not required and RNA pellets can be directly dissolved in nuclease-free water.- Dissolve the RNA pellet in 40 μl of 1x DNase buffer with 0.5 μl of DNase I, incubate it at 37 °C for 10 min, vortex the tube, and then spin down the sample briefly. Incubate the sample again at 37 °C for 10 min.
- Add 0.4 μl of 0.5 M EDTA to the RNA sample and heat the sample at 70 °C in a heat block for 10 min to inactivate the DNase I.
Data analysis
Representative data (Figure 2)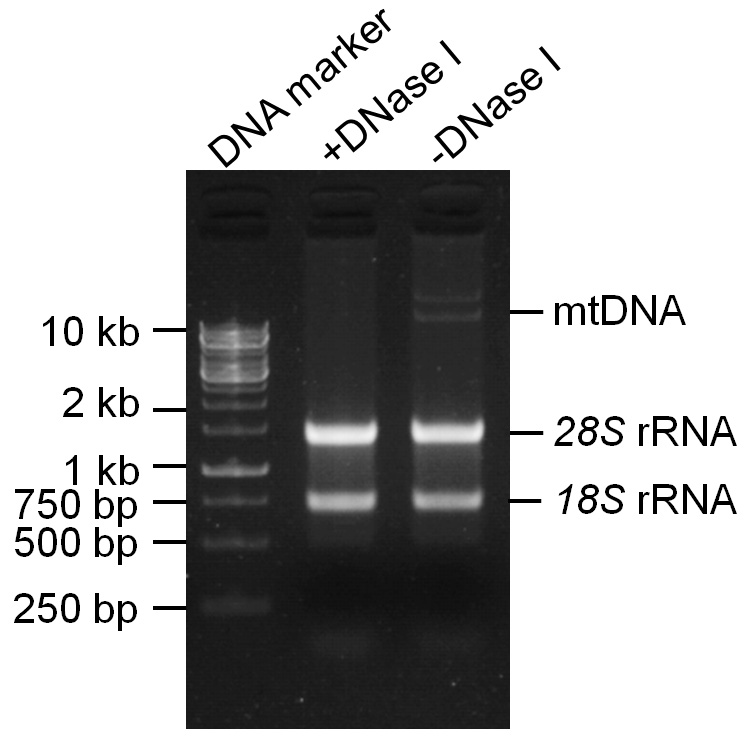
Figure 2. Agarose gel image of the isolated mitochondrial RNA, which was treated with or without DNase I. Three hundred nanograms of the isolated RNA were treated with (+DNase I) or without (-DNase I) DNase I. After DNase I inactivation, RNA samples were separated on a 1.5% agarose gel stained with ethidium bromide.
Data processing
The gel image was captured with Tanon 1600 Gel Image System (Tanon), and cropped using Photoshop.
Data analysis
No statistical analysis was performed.
Recipes
- MitoPrep buffer
0.225 M mannitol
0.075 M sucrose
20 mM HEPES (pH 7.4)
Dissolve 2.05 g of mannitol (Mw: 182.17), 1.28 g of sucrose (Mw: 342.3), and 0.2383 g of HEPES (Mw: 238.3) in ~45 ml ddH2O, adjust the pH to 7.4 with 1 M KOH, and add ddH2O to the final volume of 50 ml
Filter through a 0.22 μm filter, and store at 4 °C - 10% (w/v) SDS
Dissolve 10 g of SDS in 100 ml of ddH2O and stir until SDS is completely dissolved
Store at RT. Dilute 10% SDS to 0.6% SDS with ddH2O - 1 M Tris-HCl (pH 7.4)
Dissolve 12.114 g of Tris (Mw: 121.14) in ~80 ml of ddH2O, stir until completely dissolved
Adjust the pH to 7.4 with HCl, and make up to 100 ml with ddH2O
Filter through a 0.22 μm filter and store at RT - 0.5 M EDTA (pH 8.0)
Add 9.306 g of EDTA (Mw: 372.24) to ~40 ml of ddH2O, stir and slowly adjust the pH to 8.0 with NaOH, and make up to 50 ml with ddH2O
Filter through a 0.22 μm filter and store at 4 °C
Note: EDTA will not fully dissolve until the pH is close to 8.0. - Proteinase K (1 mg/ml)
Dissolve 1 mg of proteinase K in 1 ml of nuclease-free water, aliquot and store at -20 °C - Lysis buffer
The lysis buffer consists of 1% SDS, 10 mM EDTA and 10 mM Tris-HCl (pH 7.4)
For a 10 ml solution:
Mix 8.7 ml of nuclease-free water, 1 ml of 10% SDS, 0.2 ml of 0.5 M EDTA (pH 8.0), and 0.1 ml of 1 M Tris-HCl (pH 7.4) and make up to 10 ml with nuclease-free water - 1x DNase buffer
Dilute 10x DNase buffer (with MgCl2) to 1x with nuclease-free water - 75% ethanol (v/v)
For a 100 ml solution, add 25 ml of nuclease-free water to 75 ml of ethanol
Mix thoroughly and store at 4 °C
Acknowledgments
This work was supported by Grant 2017YFA0504600 from the Priority Research Program of the Ministry of Science and Technology of China, by Grants 31371439 and 91649103 from the National Natural Science Foundation of the People’s Republic of China, and by funds from the Ministry of Education of the People’s Republic of China 1000 Talents Youth Program. This protocol was modified from Huang et al. (2018) and Liu et al. (2017).
Competing interests
The authors declare no conflicts of interest with the contents of this article.
References
- Bruni, F., Gramegna, P., Oliveira, J. M., Lightowlers, R. N. and Chrzanowska-Lightowlers, Z. M. (2013). REXO2 is an oligoribonuclease active in human mitochondria. PLoS One 8(5): e64670.
- Cheng, Y., Liu, P., Zheng, Q., Gao, G., Yuan, J., Wang, P., Huang, J., Xie, L., Lu, X., Tong, T., Chen, J., Lu, Z., Guan, J. and Wang, G. (2018). Mitochondrial trafficking and processing of telomerase RNA TERC. Cell Rep 24(10): 2589-2595.
- Entelis, N., Kolesnikova, O., Kazakova, H., Brandina, I., Kamenski, P., Martin, R. P. and Tarassov, I. (2002). Import of nuclear encoded RNAs into yeast and human mitochondria: experimental approaches and possible biomedical applications. Genet Eng (N Y) 24: 191-213.
- Geiger, J. and Dalgaard, L. T. (2018). Isolation and analysis of mitochondrial small RNAs from rat liver tissue and HepG2 cells. Methods Mol Biol 1782: 337-350.
- Hällberg, B. M. and Larsson, N. G. (2014). Making proteins in the powerhouse. Cell Metab 20(2): 226-240.
- Huang, J., Liu, P. and Wang, G. (2018). Regulation of mitochondrion-associated cytosolic ribosomes by mammalian mitochondrial ribonuclease T2 (RNASET2). J Biol Chem 293(51): 19633-19644.
- Huang, J. and Wang, G. (2019). Organelle-associated rRNA Degradation. Bio-protocol 9(11): e3255.
- Kellems, R. E. and Butow, R. A. (1972). Cytoplasmic-type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J Biol Chem 247(24): 8043-8050.
- Levy, S., Allerston, C. K., Liveanu, V., Habib, M. R., Gileadi, O. and Schuster, G. (2016). Identification of LACTB2, a metallo-β-lactamase protein, as a human mitochondrial endoribonuclease. Nucleic Acids Res 44(4): 1813-1832.
- Liu, P., Huang, J., Zheng, Q., Xie, L., Lu, X., Jin, J. and Wang, G. (2017). Mammalian mitochondrial RNAs are degraded in the mitochondrial intermembrane space by RNASET2. Protein Cell 8(10): 735-749.
- Mager-Heckel, A. M., Entelis, N., Brandina, I., Kamenski, P., Krasheninnikov, I. A., Martin, R. P. and Tarassov, I. (2007). The analysis of tRNA import into mammalian mitochondria. Methods Mol Biol 372: 235-253.
- Mercer, T. R., Neph, S., Dinger, M. E., Crawford, J., Smith, M. A., Shearwood, A. M., Haugen, E., Bracken, C. P., Rackham, O., Stamatoyannopoulos, J. A., Filipovska, A. and Mattick, J. S. (2011). The human mitochondrial transcriptome. Cell 146(4): 645-658.
- Rorbach, J., Nicholls, T. J. and Minczuk, M. (2011). PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res 39(17): 7750-7763.
- Sidova, M., Tomankova, S., Abaffy, P., Kubista, M. and Sindelka, R. (2015). Effects of post-mortem and physical degradation on RNA integrity and quality. Biomol Detect Quantif 5: 3-9.
- Sripada, L., Tomar, D., Prajapati, P., Singh, R., Singh, A. K. and Singh, R. (2012). Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7(9): e44873.
- Wang, G., Chen, H. W., Oktay, Y., Zhang, J., Allen, E. L., Smith, G. M., Fan, K. C., Hong, J. S., French, S. W., McCaffery, J. M., Lightowlers, R. N., Morse, H. C., 3rd, Koehler, C. M. and Teitell, M. A. (2010). PNPASE regulates RNA import into mitochondria. Cell 142(3): 456-467.
- Williams, C. C., Jan, C. H. and Weissman, J. S. (2014). Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346(6210): 748-751.
- Zhang, X., Zuo, X., Yang, B., Li, Z., Xue, Y., Zhou, Y., Huang, J., Zhao, X., Zhou, J., Yan, Y., Zhang, H., Guo, P., Sun, H., Guo, L., Zhang, Y. and Fu, X. D. (2014). MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158(3): 607-619.
- Zheng, Q., Liu, P., Gao, G., Yuan, J., Wang, P., Huang, J., Xie, L., Lu, X., Di, F., Tong, T., Chen, J., Lu, Z., Guan, J. and Wang, G. (2019). Mitochondrion-processed TERC regulates senescence without affecting telomerase activities. Protein Cell doi: 10.1007/s13238-019-0612-5.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Huang, J. and Wang, G. (2019). Improved Mammalian Mitochondrial RNA Isolation. Bio-protocol 9(11): e3247. DOI: 10.21769/BioProtoc.3247.
- Huang, J., Liu, P. and Wang, G. (2018). Regulation of mitochondrion-associated cytosolic ribosomes by mammalian mitochondrial ribonuclease T2 (RNASET2). J Biol Chem 293(51): 19633-19644.
Category
Molecular Biology > RNA > RNA extraction
Cell Biology > Organelle isolation > Mitochondria
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










