- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of the Humidity Effect on HR by Ion Leakage Assay
Published: Vol 9, Iss 7, Apr 5, 2019 DOI: 10.21769/BioProtoc.3203 Views: 6105
Reviewed by: Zhibing LaiHedwin Kitdorlang DkharAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Maize Seedlings Colonization with Serendipita indica and Its Colonization Efficiency Analysis
Om Prakash Narayan [...] Atul Kumar Johri
Oct 20, 2023 2266 Views

Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries
Piao Yang [...] Ye Xia
Oct 20, 2023 2761 Views
Abstract
We describe a protocol to measure the contribution of humidity on cell death during the effector-triggered immunity (ETI), the plant immune response triggered by the recognition of pathogen effectors by plant resistance genes. This protocol quantifies tissue cell death by measuring ion leakage due to loss of membrane integrity during the hypersensitive response (HR), the ETI-associated cell death. The method is simple and short enough to handle many biological replicates, which improves the power of test of statistical significance. The protocol is easily applicable to other environmental cues, such as light and temperature, or treatment with chemicals.
Keywords: ETIBackground
Environmental cues are important factors in determining the outcome of host-pathogen interactions. The disease triangle paradigm requires, among all, a favorable environment for disease to develop (Francl, 2001; Scholthof, 2007). High humidity, for example, represses HR development and negatively regulates ETI (Zhou et al., 2004; Xin et al., 2016; Mwimba et al., 2018). It is, thus, important to quantify the effect of environments on host-pathogen interactions.
Cell death by HR during ETI is often quantified using a time-course measurement of ion leakage from infected tissue to the aqueous solution surrounding the tissues (Hatsugai and Katagiri, 2018). However, when interested in assessing the contribution of the environment to the HR development, it is necessary for tissue to remain in the environment being studied until HR has developed.
In this protocol, we subject infected tissue to 50% RH or 90% RH for 36 h before conductivity is measured. Also, we have adjusted the dosage of the pathogen to OD600nm = 0.002 to delay the onset of HR and maximize the effect of the environment on HR development. The method adapted here was originally designed to measure cell death in senescing leaves (Woo et al., 2001). Leaves were immersed in 400 mM mannitol, and conductivity was expressed as percentage of the ratio of conductivity measurement before and after boiling. In this modified method, we use tissue of equal size (leaf disc), which make reporting percent ion leakage optional. This protocol was used in our study (Mwimba et al., 2018) and can be applied to other environmental cues or to chemical treatments.
Materials and Reagents
- 200 µl and 1,000 µl pipet tips
- 1.5 ml tube (Eppendorf, catalog number: 022363204)
- 15 ml sterilized tubes (VWR, catalog number: 89039-664)
- 50 ml sterilized tubes (VWR, catalog number: 89039-656)
- 1 ml needle-less syringes for bacterial inoculation (BD, catalog number: 309659)
- Kimwipes (Kimberly-Clarck, catalog number: 34120)
- Pseudomonas syringae pv. tomato DC3000 expressing the AvrRpt2 effector (Pst AvrRpt2)
- dH2O
- MgCl2 (Sigma, catalog number: M8266) or MgSO4 (Sigma, catalog number: M7506)
- Mannitol (Sigma, catalog number: M4125)
- Agar A (Bio Basic, catalog number: FB0010)
- Proteose peptone (BD Biosciences, catalog number: 211684)
- K2HPO4·3H2O (Fisher scientific, CAS: 16788-57-1)
- 1 M MgSO4 (VWR, catalog number: 0338-500G)
- 80% Glycerol (Sigma, catalog number: G5516)
- King’s B agar (see Recipes) plate with kanamycin (50 µg/ml) and rifampicin (25 µg/ml) antibiotic selection
- 400 mM mannitol solution (see Recipes)
Equipment
- 1 L beaker
- Shaker (Lab Companion, SI-600R)
- Water bath (Precision Scientific, Model 25)
- Growth chambers (Percival, AR36L3)
- Pipets (Eppendorf, 20-200 µl and 100-1,000 µl)
- Spectrophotometer (Amersham Biosciences, model: Ultraspec 2100 pro)
- Electro-conductivity meter (Thermo Scientific, model: Orion Star A322)
- Puncher and sampling tool (Electron Microscopy Sciences, model: EMS Rapid-core 6.0, catalog number: 69039-60)
Software
- Microsoft Excel
- Graph prism, or R
Procedure
- Grow 12 plants per genotype on soil for 3 weeks under 22 °C, 50% RH 12 h light/12 h dark condition.
- In a 15 ml tube, prepare Pst AvrRpt2 (OD600nm = 0.002) in 10 mM MgCl2 (or MgSO4).
- Using a 1 ml needle-less syringe, pressure-infiltrate 3 fully expanded leaves (leaves 4, 5 and 6)
per plants with the Pst AvrRpt2.
Note: It is much easier to infiltrate from the abaxial side of the leaf. - Dry excess of Pst AvrRpt2 inoculum gently using Kimwipes.
- Move plants to chambers set to conditions being compared. Keep 6 plants per genotype under 50% RH 22 °C 12 h light/12 h dark and the other 6 plants per genotype 90% RH 22 °C 12 h light/12 h dark conditions for 36 h.
Note: We use Pst AvrRpt2 at OD600nm = 0.002 to capture the effect of the environment on the developing ETI. At this inoculant, the hypersensitive response becomes visible to the eye at around 24 hpi in plants under 50% RH. - Collect dH2O in a 1 L beaker and confirm that the conductivity is below 1.5 µS/cm.
Note: High dH2O conductivity generates high noise and can result in failure of the experiment. - Using the EMS Rapid-core 6.0 puncher, collect 3 leaf discs per plant (1 disc per leaf) in a 50 ml tube (i.e., 6 tubes per genotype).
- Label each tube individually if interested to report data as a percentage (see Step 15)
- Add 30 ml dH2O to each tube and rinse discs by inverting tube 3 times.
- Using dH2O, prepare 100 ml of 400 mM mannitol (7.3 g mannitol in 100 ml dH2O) per genotype.
Note: Confirm ion measurement of the 400 mM mannitol is not greater than 3 µS/cm. - For each tube containing leaf discs, replace all dH2O with 6 ml 400 mM mannitol.
- Make a blank by adding 6 ml 400 mM mannitol to an empty 50 ml tube for measurement correction during data analysis
- Shake tubes at 100 rpm for 2 h at room temperature.
- Using the Thermo Scientific Orion Star A322 electro-conductivity meter, measure initial ion leakage (see Figure 1) and proceed to data analysis.
Note: When measuring, be careful not to hurt leaf discs or move discs across tubes during measurement. - (Optional Step) If presenting data as percent ion leakage.
- For 10 min, boil samples by moving tubes to a water bath set at 100 °C.
- Cool tubes to RT.
- Measure total ion leakage and proceed to data analysis.

Figure 1. Picture describing ion leakage measurement using the Thermo Scientific Orion Star A322 Electro-conductivity meter
Data analysis
- Present data as corrected conductivity values or percent ratio of corrected initial to corrected total conductivity (see Figure 2).
Corrected conductivity = Measured conductivity - Conductivity of blank.
Percent ratio =100 x (Initial corrected conductivity/Total corrected conductivity)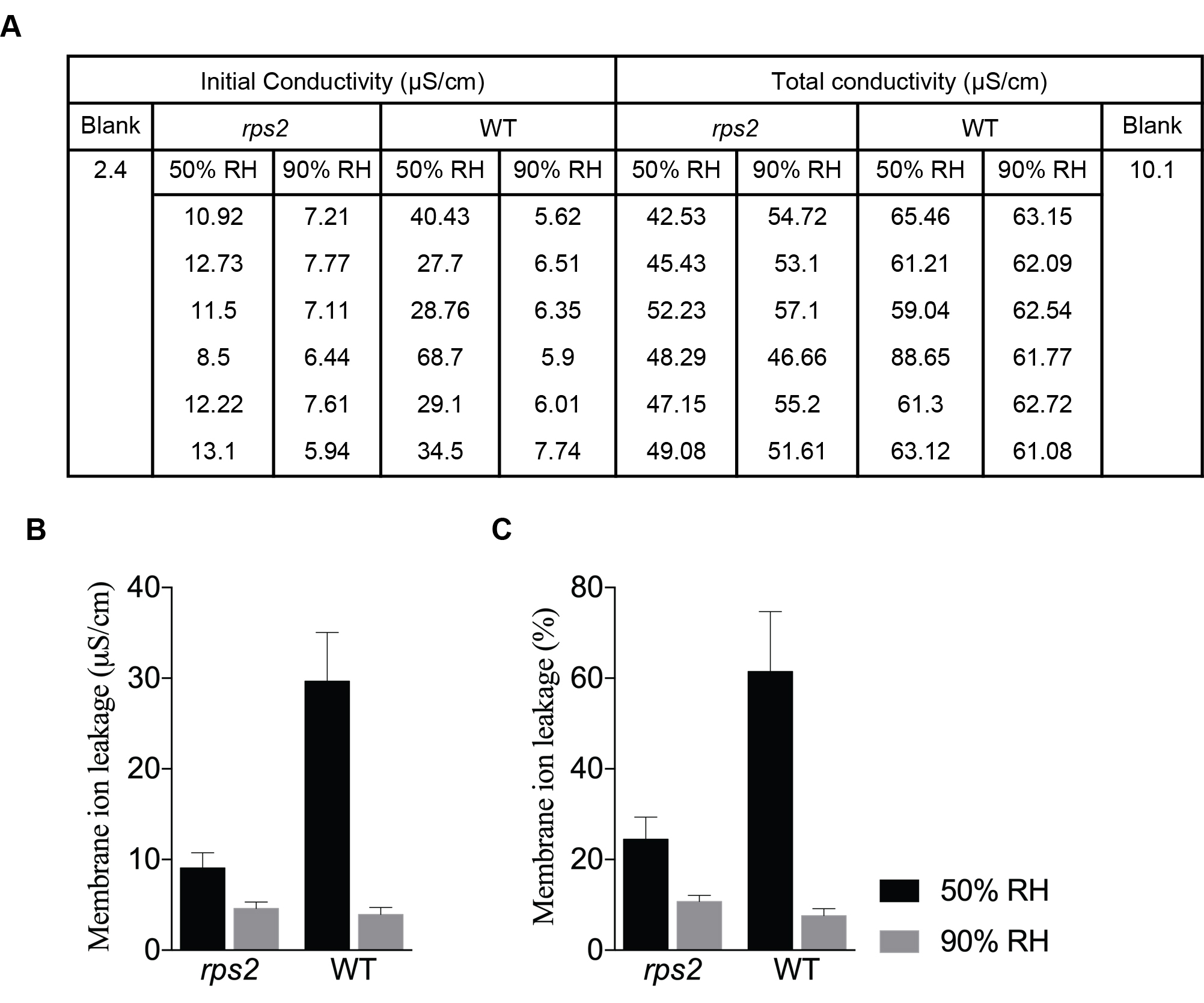
Figure 2. High humidity suppresses the hypersensitive response. A. Conductivity values measured 36 hpi and after boiling. Plants were moved to 50% RH or 90% RH after inoculation with Pst AvrRpt2 OD600nm = 0.002. B and C. Quantification of the hypersensitive response, described in (A), was graphed as membrane ion leakage measurement (B) and as percent membrane ion leakage (C). Data are mean ± S.D. - Repeat the experiment at least three independent times for data reproducibility.
- For statistical analysis, use two-way ANOVA. The three experimental replicates can be combined using linear mixed-effect models.
Recipes
- King’s B agar (1 L)
15 g Agar A
20 g Proteose peptone
2 g K2HPO4·3H2O
Autoclave for 20 min at 120 °C then add 6.1 ml of autoclaved 1 M MgSO4, 18.8 ml of autoclaved 80% Glycerol - 400 mM mannitol solution (1 L)
Add 72.88 g mannitol to 500 ml dH2O
Stir to dissolve mannitol
Add dH2O to bring volume to 1 L
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) (1R01-GM099839-01, 2R01-GM069594-09, and 5R35-GM118036) and by the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (GBMF3032). We have adapted this protocol from Woo et al. (2001).
Competing interests
The authors have no conflicts of interest or competing interests.
References
- Francl, L. J. (2001). The disease triangel: a plant pathological paradigm revisited. The Plant Health Instructor. Doi: 10.1094/PHI-T-2001-0517-01.
- Hatsugai, N. and Katagiri, F. (2018). Quantification of plant cell death by electrolyte leakage assay. Bio-protocol 8(5): e2758.
- Mwimba, M., Karapetyan, S., Liu, L., Marques, J., McGinnis, E. M., Buchler, N. E. and Dong, X. (2018). Daily humidity oscillation regulates the circadian clock to influence plant physiology. Nat Commun 9(1): 4290.
- Scholthof, K. B. (2007). The disease triangle: pathogens, the environment and society. Nat Rev Microbiol 5(2): 152-156.
- Woo, H. R., Chung, K. M., Park, J. H., Oh, S. A., Ahn, T., Hong, S. H., Jang, S. K. and Nam, H. G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13(8): 1779-1790.
- Xin, X. F., Nomura, K., Aung, K., Velasquez, A. C., Yao, J., Boutrot, F., Chang, J. H., Zipfel, C. and He, S. Y. (2016). Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539(7630): 524-529.
- Zhou, F., Menke, F. L., Yoshioka, K., Moder, W., Shirano, Y. and Klessig, D. F. (2004). High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J 39(6): 920-932.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mwimba, M. and Dong, X. (2019). Quantification of the Humidity Effect on HR by Ion Leakage Assay. Bio-protocol 9(7): e3203. DOI: 10.21769/BioProtoc.3203.
Category
Plant Science > Plant physiology > Biotic stress
Developmental Biology > Cell signaling > Stress response
Biochemistry > Other compound > Ion
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









