- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Flip-flop Mediated Conditional Gene Inactivation in Drosophila
Published: Vol 9, Iss 3, Feb 5, 2019 DOI: 10.21769/BioProtoc.3157 Views: 7908
Reviewed by: Miao HeRajesh D GunageAbhijit Kale

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

CRISPR-Cas9 Mediated Genome Editing in Drosophila
Ping Peng [...] Jian-Quan Ni
Jan 20, 2019 11129 Views

Library Preparation for Genome-Wide DNA Methylation Profiling
Fei-Man Hsu [...] Pao-Yang Chen
Nov 5, 2025 1575 Views
Abstract
Mosaic analysis in Drosophila, an important tool to assess cellular phenotypes of mutants in an otherwise heterozygous background, relies on mitosis. Hence, it cannot be used to inactivate gene function in mitotically inactive, terminally differentiated cells such as neurons. To address this issue, we developed “Flip-flop”, a novel, Flippase-dependent in vivo cassette-inversion method that functions independent of mitosis, and therefore can be used for gene inactivation in both mitotic as well as post-mitotic cells. This method allows tagging protein-coding genes with EGFP and generates mutant cells that are marked with mCherry upon cassette inversion. Here, we describe protocols for generation and validation of fly lines that can be used for conditional gene inactivation in mitotic as well as post-mitotic cells. We provide typical examples of Flip-flop mediated mosaic analysis in SNF4Aγ and Trim9. Use of Flip-flop mediated functional analysis will permit a detailed investigation of the role of genes previously recalcitrant to mosaic analysis.
Keywords: DrosophilaBackground
We developed a Flippase/Flippase recognition target (FLP/FRT) based method for mosaic analysis and conditional gene inactivation in Drosophila that is independent of mitotic separation of mutant alleles. The development and early testing of the method are described in Nagarkar-Jaiswal et al., 2017. Here, we present a protocol for establishment and evaluation of the fly stocks that can be used for functional annotation of genes in a tissue-specific manner throughout the development of Drosophila. The method, called “Flip-flop”, is built upon the versatile Minos mediated integration cassette (MiMIC) platform (Venken et al., 2011; Nagarkar-Jaiswal et al., 2015a and 2015b) and involves genomic integration of the “Flip-flop cassette” that contains two modules: a protein-trap (PT) module and a gene-trap (GT) module (Figure 1A). The PT-module tags the gene of interest with EGFP at the endogenous locus, whereas the GT module facilitates generation of truncation mutants that express mCherry under the control of the natural regulatory elements of the gene. Once the cassette is inserted in a coding intron in the PT orientation, it can be reversed into the opposite GT-orientation in vivo with the help of FLP. The inversion marks the mutant cells, as they express mCherry under the endogenous gene’s control, and helps distinguish them from the rest of the tissue expressing EGFP tagged protein (Figure 1B). There are several advantages of Flip-flop over the traditional method MARCM (Mosaic Analysis with a Repressible Cell Marker; Lee and Luo, 1999): First, the control cells and the mutant cells are marked with EGFP and mCherry respectively, and the reporters are expressed at the endogenous level. This removes the need for introducing additional fluorescent reporters and GAL4 drivers required in MARCM. Second, Flip-flop circumvents an often overlooked problem of MARCM, wherein secondary mutations distal to the primary mutation of interest also become homozygous in the mutant cells. Flip-flop involves a local change within the loci of the gene of interest and therefore creates mutant cells with a specific, clearly-marked mutation. Hence, there is no general need to validate Flip-flop experiments in order to test whether the phenotype is due to the mutation within the gene of interest by performing a rescue experiment. Third, unlike MARCM, this method does not rely on cell division and can be used for conditional gene inactivation in post-mitotic cells such as neurons. Moreover, the PT orientation of Flip-flop reveals the natural expression pattern and protein localization of the gene in which they are inserted. Together, these advantages will allow Flip-flop users to rapidly develop reagents necessary for conditional inactivation of genes that permit functional analysis at unprecedented detail. 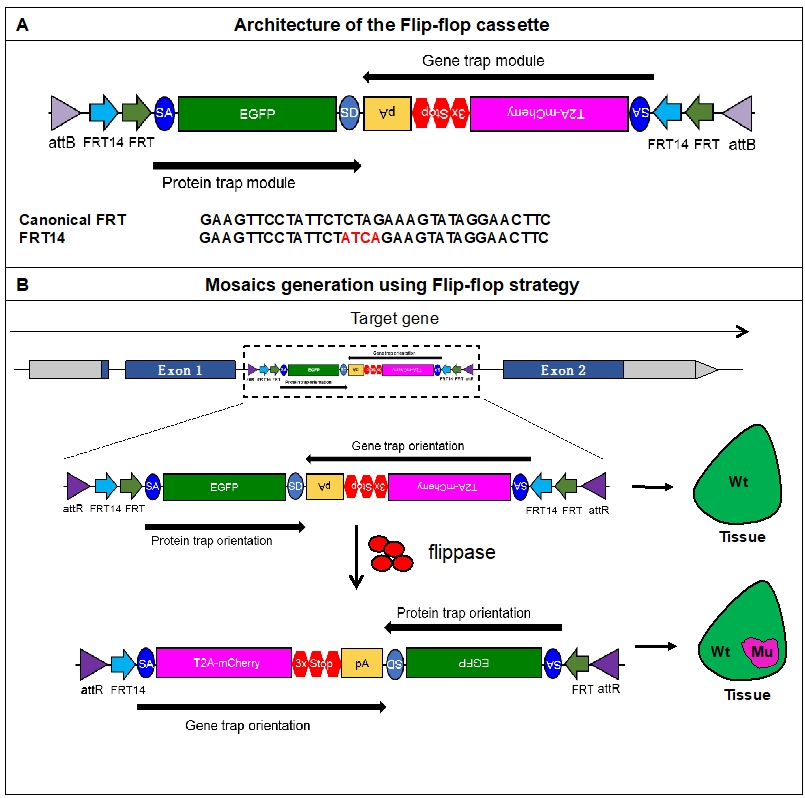
Figure 1. Mosaic generation using the Flip-flop cassette. A. The architecture of the Flip-flop cassette. The cassette consists of two independent modules (PT and GT), that are oriented in opposite orientations. The PT module contains a splice acceptor (SA), followed by an EGFP tag and a splice donor (SD). The GT module contains an SA sequence, followed by the T2A peptide coding sequence (which will induce a translational skip), the mCherry coding region, stop codons in all three coding frames, and an SV40 polyA transcriptional termination signal. Given the opposite orientation of both modules, only one of the SA sequence will be active with respect to the recipient gene. The two modules are nested within a pair of FRT and FRT14 inverted repeats, forming an FLP-responsive FLEx switch. Finally, the entire cassette is flanked by two inverted attB sequences that permit phiC31 mediated recombination-mediated cassette exchange (RMCE) between the Flip-flop cassette and pre-existing MiMIC elements. A comparison of the FRT and FRT14 sequence is shown below. The FRT14 sequence varies from the canonical FRT sequence at the residues highlighted in red. B. Schematic showing the inversion of a PT-oriented Flip-flop cassette, inserted into the coding intron of a hypothetical gene. Upon FLP-expression, the “Flip-excision” (FLEx) switch undergoes two recombination events: (1) recombination between the two FRT sites or between the two FRT14 sites leads to cassette inversion that is followed by (2) excision of either the pair of FRT sites or the pair of FRT14 sites. The pair that is excised during the second step is dependent on the pair that underwent recombination in the first step. e.g., if the first recombination occurred between two FRT sites, the FRT14 sites will be turned into the same orientation and will recombine in the second step. This will remove one of the FRT and one of the FRT14 sites. Conversely, if the FRT14 sites recombined in the first step, the FRT sites will recombine in the second step, and once again remove one of the FRT and one of the FRT14 sites. But, following the second recombination step, the remaining unpaired FRT and FRT14 sites are not able to recombine, and the cassette will be locked in the GT orientation. Thus, the initial PT orientation allows the gene to be tracked by EGFP-tagged protein expression in tissues. FLP activity inverts the Flip-flop cassette in random cells, generating a mosaic tissue consisting of cells that did not undergo the flip and are still expressing the EGFP-tagged protein and cells that inverted the Flip-flop cassette into the GT orientation, which is marked by mCherry expression. (Adapted from Nagarkar-Jaiswal et al., 2017.)
Below, we describe the procedure to develop Flip-flop stocks from two existing MiMIC lines through embryonic injection and transgenesis, as well as molecular confirmation of stocks using PCR. We have also described some experimental strategies that can be used to evaluate Flip-flop in different cell types at different stages of development.
Materials and Reagents
- Consumables
- Small sieve for washing embryos (Nylon mesh 150, pore size about 100 microns)
- Artist’s brush (0 number painting brush)
- Double sided tape
- Slides and coverslips
- Plasmids
pFlip-flop-P0 for phase 0
pFlip-flop-P1 for phase 1
pFlip-flop-P2 for phase 2
p3xP3-EGFP.vas-int.NLS
PhiC31 integrase expressing vector
Note: All the constructs can be obtained from the Drosophila Genomics Resource Center (DGRC). - Fly stocks
Gene specific MiMIC insertion (Nagarkar-Jaiswal et al., 2015)- Stock number: 53104 Genotype: y1w*;Mi{MIC}SNF4AγMI09417/TM3,Sb1Ser1 (Gene: SNF4Agamma)
- Stock number: 58593 Gentotype: y1w*;Mi{MIC}Trim9MI12525 (Gene: Trim9)
Additional stocks for genetic manipulation and evaluation- y1w67c23;In(2LR)Gla, wgGla−1/SM6a
- y1w67c23;D/TM6b, Hu, Tb
- w[*];L[2] Pin[1]/CyO,P{w[+mC]=GAL4-Kr.C}DC3,P{w[+mC]=UAS-GFP.S65T}DC7
- y[1]w[*];D*gl[3]/TM3,P{w[+mC]=GAL4-Kr.C}DC2,P{w[+mC]=UAS-GFP.S65T}DC10, Sb[1]
- P{ry[+t7.2]=hsFLP}12,y[1]w[*];sna[Sco]/CyO
- P{ry[+t7.2]=hsFLP}12,y[1]w[*];Dr e/TM3,Sb
- y[d2]w[1118] P{ry[+t7.2]=ey-FLP.N}2
- P{w[+mC]=tubP-GAL80[ts]},w[*]/FM7a;P{w[+mC]=UAS-FLP.D}JD2
- y[1]w[*];P{w[+mC]=UAS-FLP.D}JD1
- y[1]w[1118];P{y[+t7.7] w[+mC]=nSyb-GAL4.P}attP2
- PCR Primers
Tag-specific primers:
(1) EGFP-TAG-Rev: GAGTACAACTTCAACAGCCAC
(2) mCherry-TAG-Fwd: GGCCACCCTTGGTCACCT
MiMIC specific primers:
(1) MiL-F: GCGTAAGCTACCTTAATCTCAAGAAGAG
(2) MiL-R: CGCGGCGTAATGTGATTTACTATCATAC - Reagents
- Yeast paste
- Bleach (50% Clorox; final concentration: 2.5% hypochlorite), freshly prepared
- Halocarbon oil 700 (Sigma, catalog number: H8898)
- PureLinkTM Genomic DNA Mini Kit (Invitrogen, catalog number: K182001)
- GoTaq® Green Master Mix (Promega, catalog number: M712)
- Formaldehyde (37% w/v, Sigma-Aldrich, catalog number: 252549)
- Normal goat serum (Jackson ImmunoResearch Labs, catalog number: 005-000-001)
- PBS with 0.2% (v/v) Triton X-100
- Anti-GFP antibody (Abcam, catalog number: ab13970)
- Anti-mCherry antibody (Clontech, catalog number: 632496)
- Secondary antibodies (Anti-Chicken Alexa 488, Jackson ImmunoResearch Labs, catalog number: 703-545-155; Anti-Rabbit Cy3, Jackson ImmunoResearch Labs, catalog number: 111-585-003)
- Agar
- Sugar
- Welch's grape juice concentrate
- Ethanol
- Glacial acetic acid
- Sucrose
- Grape juice agar plates (see Recipes)
- 10% sucrose agar plates (see Recipes)
Equipment
- Fly cages
- FemtoJet 4x (Eppendorf, catalog number: 5253000017)
- Femtotips (Eppendorf, catalog number: E5242952008)
- Incubator for culturing flies
- Circulating water bath (Thomas Scientific Julabo, catalog number:1181j40)
- Dissecting microscope (Stemi 305 Microscope) (Zeiss, catalog number: 435063-9010-100)
- Fine dissection tools:
Scissors (Fine Science Tools, catalog number: 16143-12)
Forceps (Fine Science Tools, catalog number: 11413-12) - Microwave (LG, catalog number: MS3042GM)
Procedure
- Choosing the MiMIC and determining the appropriate phase of the MiMIC lines
- Publicly available MiMIC lines are listed in Bloomington Drosophila Stock Center (BDSC) database and can be accessed from FlyBase (http://Flybase.org) or at the stock center website (https://bdsc.indiana.edu).
- Choose a MiMIC insertion that tags your gene of interest, preferably one that tags all of the transcriptional isoforms of the gene (called “gold” MiMICs in Nagarkar-Jaiswal et al., 2015). As an example, in this protocol, we are using MiMIC lines that tag the genes SNF4Agamma and Trim9.
- Using RMCE resources page (http://flypush.imgen.bcm.tmc.edu/pscreen/rmce/) from the Bellen lab at Baylor College of Medicine, determine the phase for the chosen MiMIC line. The proper functioning of the Flip-flop cassette is dependent on the in-frame translation of the Flip-flop cassette with the preceding exon. The in-frame translation is determined by the last codon of the preceding exon (preceding codon) and can result in one of the three phases of translation for Flip-flop: Phase 0 or Phase 1 or Phase 2. Choose the corresponding plasmid donor for the Flip-flop as dictated by the MiMIC insertion. Also, determine the relative orientation of the MiMIC element with that of the recipient gene. This information is necessary for the molecular validation of Flip-flop cassette insertion using PCR.
- Generation of Flip-flop fly lines
In this section, we describe the generation of the Flip-flop alleles by co-injecting the Flip-flop cassettes with the PhiC31 integrase plasmid into embryos carrying the MiMIC insertion.- Using a stock of flies carrying gene specific MiMIC (yellow+) inserted in a coding intron [F0], set up a small fly cage (~150-300 adult flies, about 50-100 males and 100-200 females that are less than 15 days old) and collect 0.5-1 h old embryos of on grape juice plates supplemented with yeast paste.
- Prepare embryos [F1] for microinjection as described in Kiehart et al. (2007). Briefly, collect the embryos from the grape juice plate and wash with water. Resuspend them in 50% bleach and stir continuously for about 2 min. Wash off the bleach and align the embryos in a line on a small piece of grape juice agar. Stick the embryos on a coverslip with double-sided tape by gently lowering the coverslip onto the embryos. Cover them with Halocarbon oil 700.
- Co-inject the phase-specific Flip-flop plasmid (concentration 400 ng/µl) and PhiC31 integrase expressing vector (concentration 300 ng/µl) along with the food color into embryos using the Femtotips and FemtoJet 4x as described in Kiehart et al., 2000.
- Let the embryos develop at 18 °C.
- Collect individual male/female adults from F1, and cross them to appropriate balancer stock depending on the chromosomal location of the MiMIC insertion. From these crosses, screen for flies that have lost the MiMIC-yellow marker (yellow-) indicating successful RMCE event [F2]. Self-cross transgenic F2 flies and establish Flip-flop stocks.
- Confirmation of orientation of the Flip-flop cassette
The Flip-flop cassette is flanked by two inverted attB that allow replacement of the MiMIC core located between two inverted attP sites (Figure 2). Since recombination can occur between any of two attP and attB sites, the Flip-flop cassette can be inserted in either a PT or a GT orientation (Figure 2). In this section, we will determine the orientation of the Flip-flop transgenic line.- From the stock of Flip-flop flies that was established in the previous section, extract genomic DNA from 8-10 flies using the PureLinkTM Genomic DNA Mini Kit (Invitrogen).
- Perform four PCR reactions for each Flip-flop line to confirm the orientation of the Flip-flop cassette (protein trap versus gene trap): (1) First PCR reaction with MiL-F and EGFP-TAG-Rev primers (2) second reaction with Mil-F and mCherry-TAG-Fwd primers, (3) third reaction with MiL-R with EGFP-TAG-Rev primers and (4) the fourth reaction with MiL-R and mCherry-TAG-Fwd primers.
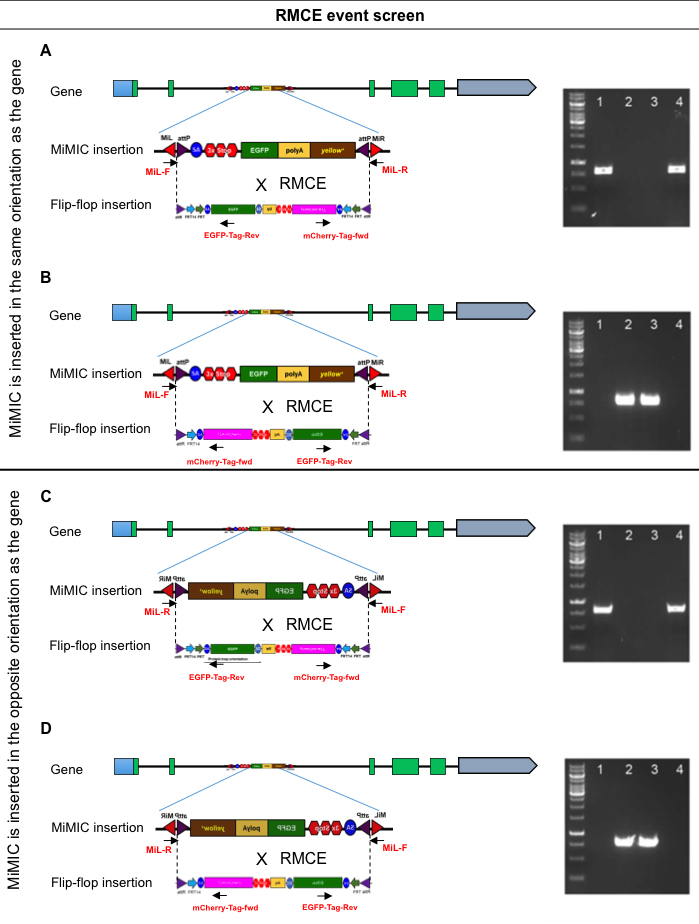
Figure 2. RMCE event screening. By knowing of the orientation of the MiMIC insertion with respect to the gene using Step A3, the PCR products as shown in (A), (B), (C) and (D) can be used to determine the orientation of the Flip-flop cassette with respect to the gene: when the MiMIC cassette is inserted in the same orientation as the gene (A) and (B), and when MiMIC is inserted in the opposite orientation of the gene (C) and (D). Gel images on the left hand side show amplification in different PCR reactions that distinguish between integration of Flip-flop in PT or GT orientations: (1) First PCR reaction with MiL-F and EGFP-TAG-Rev primers (2) second reaction with Mil-F and mCherry-TAG-Fwd primers, (3) third reaction with MiL-R with EGFP-TAG-Rev primers and (4) the fourth reaction with MiL-R and mCherry-TAG-Fwd primers. When the MiMIC insertion is in the same orientation as the gene (SNF4Aγ), a positive amplification in reactions 1 and 4 will confirm the PT orientation (A), while positive bands in 2 and 3 will confirm the GT orientation (B). If the MiMIC insertion and the gene are in the opposite orientation (Trim9), positive reactions in 2 and 3 will confirm the PT insertion (C) and 1 and 4 will confirm the GT insertion (D). - Induction of Flip-flop cassette inversion to facilitate conditional knock-down
In this section, we provide generic schemes for generating experimental animals that can be used for Flip-flop mediated mosaic analysis in different tissues using different sources of FLPs (Figure 3). We refer to the gene of interest as “gene” in these schemes. Consequently, genenull is used to refer to a strong loss of function allele that is derived from a different source other than the Flip-flop stock. Flip-flop stocks are indicated with geneEGFP-T2A-mCherry- indicating that they are in the PT orientation. We also provide selected examples of results of mosaic tissue generated using Flip-flop in mitotic as well as post-mitotic cells from our previous study. We track the formation of mosaics using genetically encoded fluorescent markers that are expressed from the genomic location of the corresponding genes.
For mosaic analysis using Flip-flop, we recommend using a combination of Flip-flop and a strong loss of function allele (genenull) in order to prevent the confounding effects of secondary mutations that may be present in the Flip-flop line. This is standard practice in Drosophila genetics, as an independently derived allele of the gene, it is unlikely to also have the same secondary mutations as Flip-flop. We also recommend the use of fluorescent balancers, such as TM3,Kr-GAL4 UAS-GFP or TM3,P{w[+mC]=sChFP}3,Sb[1] which permit easy identification of non-balancer-carrying animals (inferred as geneEGFP-T2A-mCherry/genenull larvae) through the absence of GFP or mCherry signal.
Conditional gene inactivation in mitotic cells
In this section, we describe Flip-flop mediated gene inactivation in mitotically active cells in a tissue-specific manner as well as in random mosaics. We used a Flip-flop insertion in a gene encoding AMP kinase gamma subunit, SNF4Aγ, as an example. To reverse the Flip-flop insertion in SNF4Aγ in a tissue-specific fashion we used eyeless promoter driven FLP (ey-FLP). This promoter drives the expression of FLP in the presumptive eye-antenna imaginal disc as well as part of the optic lobe of the larval brain. For random mosaics generation throughout the animal’s body, we used heat-shock inducible FLP (hs-FLP) expression. The expression of FLP from this source is stochastic and induced upon a heat shock to the larvae at 37 °C.- Flip-Flop cassette inversion in eye antenna disc
- Using standard genetic crosses create appropriate fly stocks carrying a transgenic source of eyeless FLP and a severe loss of function allele and null mutants of the gene (ey-FLP/Y;genenull/CyO,Kr-GAL4 UAS-GFP or TM3,Kr-GAL4 UAS-GFP) and cross them with geneEGFP-T2A-mCherry/geneEGFP-T2A-mCherry carrying flies. A schematic of cross strategy is shown in Figure 3A. Raise these crosses at 25 °C.
- Collect larvae 48 h post-egg-laying and select non-GFP third instar larvae. Dissect the eye-antenna disc under the dissecting microscope and stain with anti-GFP and anti-mCherry antibodies as described in Nagarkar-Jaiswal et al., 2017. Briefly, fix the dissected larvae with 4% formaldehyde for 30 min, wash three times with PBS-0.2% Triton X-100 and block with 10% Normal Goat Serum for 1 h at room temperature. Incubate samples with anti-GFP and anti-mCherry at 4°C overnight. Wash samples three times with PBS-0.2% Triton X-100 and incubate with Secondary antibodies for 2 h at room temperature. Mount the sample in mounting media.
- Mosaic generation in the adult gut
- Create fly stocks hs-FLP/Y;genenull/CyO,Kr-GAL4 UAS-GFP or TM3,Kr-GAL4 UAS-GFP using conventional genetic approach and then cross them with y1w*;geneEGFP-T2A-mCherry/geneEGFP-T2A-mCherry Flip-flop flies and keep at 18 °C. A schematic of cross strategy is shown in Figure 4A. Collect appropriate progeny carrying the Flip-flop allele and the null allele (hs-FLP/y1w*;genenull/geneEGFP-T2A-mCherry) and maintain them at 18 °C for 3 days.
- Heat shock adult females on Day 3 for 2 h at 37 °C and keep them at 25 °C.
- Optional: Starvation assay or drug feeding assay could be performed to compare the effect of starvation or drug on the mutant gut cells.
- Dissect guts and stain with anti-GFP and anti-mCherry antibodies as described in Nagarkar-Jaiswal et al., 2017.
Conditional gene inactivation in mitotically inactive cells
Neurons are mitotically inactive cells. This precludes the use of traditional mosaic analysis techniques such as MARCM, which depend on mitotic separation of mutant and heterozygous cells. Application of Flip-flop, however, does not require mitotic separation of the mutant cells and therefore can be used to conditionally inactivate genes in adult neurons. To demonstrate the applicability of Flip-flop to non-mitotic cells such as neurons, we describe conditional inactivation of Trim9 in adult neurons by using neuronal driver nSyb-GAL4 and UAS-FLP (Figures 5A and 5B). We chose nSyb-GAL4 as it is a well-characterized driver of GAL4 expression in mature neurons. However, it is constitutively expressed in neurons. To achieve conditional inactivation in adult neurons and not in larval neurons, we limited the activity of nSyb-GAL4 during development using a temperature sensitive GAL80 (GAL80ts). GAL80 is a well-known inhibitor of GAL4 and can be used to suppress the expression of GAL4/UAS-driven FLP during development. Maintaining the suppressed FLP (GAL4/UAS-FLP, GAL80ts) carrying stocks at low temperature and then switching to a higher temperature in adults will restrict FLP expression at low temperature and drive the expression of FLP at high temperature. Hence, a temperature shift post-development will lead to Flip-flop cassette inversion predominantly in the adults. This approach can be used for the identification of adult neuronal functions of genes which are essential for proper development. Below, we describe a generic scheme wherein any neuronal GAL4 can be used to generate stocks that permit conditional gene inactivation in neurons during development as well as in adult brain using Flip-flop (Figure 5A).
Gene inactivation in neurons
- Create fly stocks tubP-GAL80ts;genenull/CyO,Kr-GAL4 UAS-GFP or TM3,Kr-GAL4 UAS-GFP;UAS-FLP/UAS-FLP and cross them to geneEGFP-T2A-mCherry/geneEGFP-T2A-mCherry;Neuronal-GAL4/Neuronal-GAL4 using conventional genetic approach. Maintain the crosses at 18 °C until eclosion.
- Shift the adults to 29 °C on Day 4 after eclosion. Dissect the brains on Day 4 after shifting to 29 °C and stain as described in Nagarkar-Jaiswal et al., 2017.
- Flip-Flop cassette inversion in eye antenna disc
Data analysis
Flip-flop mediated mosaic generation in the eye-antennal imaginal discs and the adult gut allows to evaluate the requirement of SNF4Aγ in eye development and in the induction of autophagy in adult guts. We also used Flip-flop to generate mutant mosaics in the adult brain. In this section, we describe the functional insights into the role of SNF4Aγ, obtained through the use of Flip-flop using methods described in Procedures A, B, and D.
- Generation of SNF4Aγ and Trim9 Flip-flop lines:
We generated SNF4Aγ and Trim9 Flip-flop lines by co-injecting Flip-flop constructs and PhiC31 integrase plasmids as described in Procedure A. We further confirmed the orientation of Flip-flop cassette by PCR assay described in Procedure C. We established biologically functional protein trapped lines for both the genes (Figure 2A and 2B), which were further used for Flip-flop assays. - SNF4Aγ is essential for proper pattern formation during eye development:
We used a severe loss of function allele SNF4AγDf along with the Flip-flop insertion in PT orientation derived from the y1w*;Mi{MIC}SNF4AγMI09417/TM3,Sb1Ser1. We crossed ey-FLP; SNF4AγDf/TM6b, Hu, Tb flies with y1w*;SNF4AγMI09417-GFP-T2A-mCherry/SNF4AγMI09417-GFP-T2A-mCherry and collected ey-FLP/y1w*;SNF4AγDf/SNF4AγMI09417-GFP-T2A-mCherry progeny. We dissected the eye-antennal discs from these animals and stained for EGFP and mCherry expression. We found the robust inversion of Flip-flop cassette in these eye discs, marked by the switch from EGFP expression in control animals to mCherry expressing mosaics in ey-FLP carrying flies (Figure 3B). Following up on the developmental inversion of Flip-flop, We also found a marked change in the arrangement of eye ommatidia and an electrical response to stimulation with light (Electroretinogram, ERG) in the adult eyes in SNF4Aγ mutant mosaics (described in Nagarkar-Jaiswal et al., 2017). - SNF4Aγ is required for autophagy induction in adult gut following starvation
We used a severe loss of function allele SNF4AγMI09417 along with the Flip-flop PT insertion derived from the same MiMIC insertion, SNF4AγMI09417-FF-EGFP-mCherry. We crossed hs-FLP/Y;SNF4AγMI09417/TM6b, Hu, Tb flies with y1w*;SNF4AγMI09417-GFP-T2A-mCherry/SNF4AγMI09417-GFP-T2A-mCherry and collected hs-FLP/y1w*;SNF4AγMI09417/SNF4AγMI09417-GFP-T2A-mCherry progeny flies. We gave heat shocks to the collected flies on Days 4 and 5 at 37 °C in an air-incubator for 2 h and starved by keeping them in vials containing 10% sucrose agar for 24 h (Figure 4B). Following starvation, we analyzed adult guts for Flip-flop inversion and induction of autophagy (Figure 4C). We found that the SNF4Aγ mutant flies fail to induce autophagy (Nagarkar-Jaiswal et al., 2017).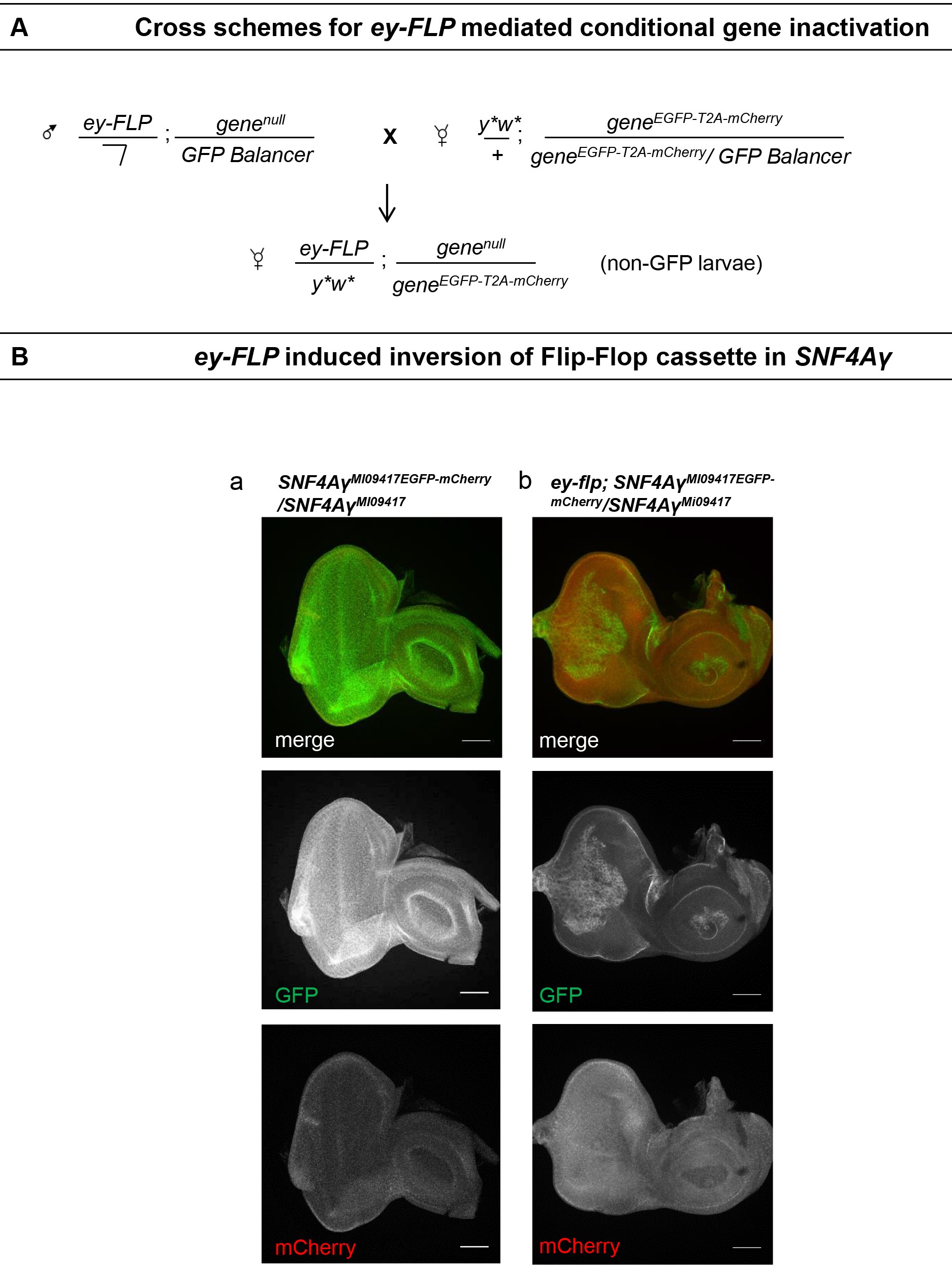
Figure 3. ey-FLP mediated conditional gene inactivation. A. Schematic showing cross strategy for Flip-flop cassette inversion in the eye-antennal disc. B. Eye-antennal discs from ey-FLP/y1w*;SNF4AγDf/SNF4AγMI09417-GFP-T2A-mCherry larvae showing loss of EGFP signal (green) and mCherry (red) following tissue-specific switch of the Flip-flop cassette with ey-FLP. Scale bars = 50 µm. (Adapted from Nagarkar-Jaiswal et al., 2017) - Flip-flop mediated loss of Trim9 in adult neurons
We used a deficiency spanning Trim9 (Trim9Df) as a loss of function allele in examining the PT insertion in Trim9. We used tubP-GAL80ts to suppress FLP-expression during development and raised Trim9MI12525EGFP-T2A-mCherry/Trim9Df;UAS-FLP/nSyb-GAL4 animals at 18 °C until four days after eclosion. Following four days of adulthood at 18 °C, the animals were shifted to 29 °C to induce FLP-expression. As expected, immunostaining the brain of these animals shows mCherry-expressing neuronal cells in which the cassette inversion has occurred.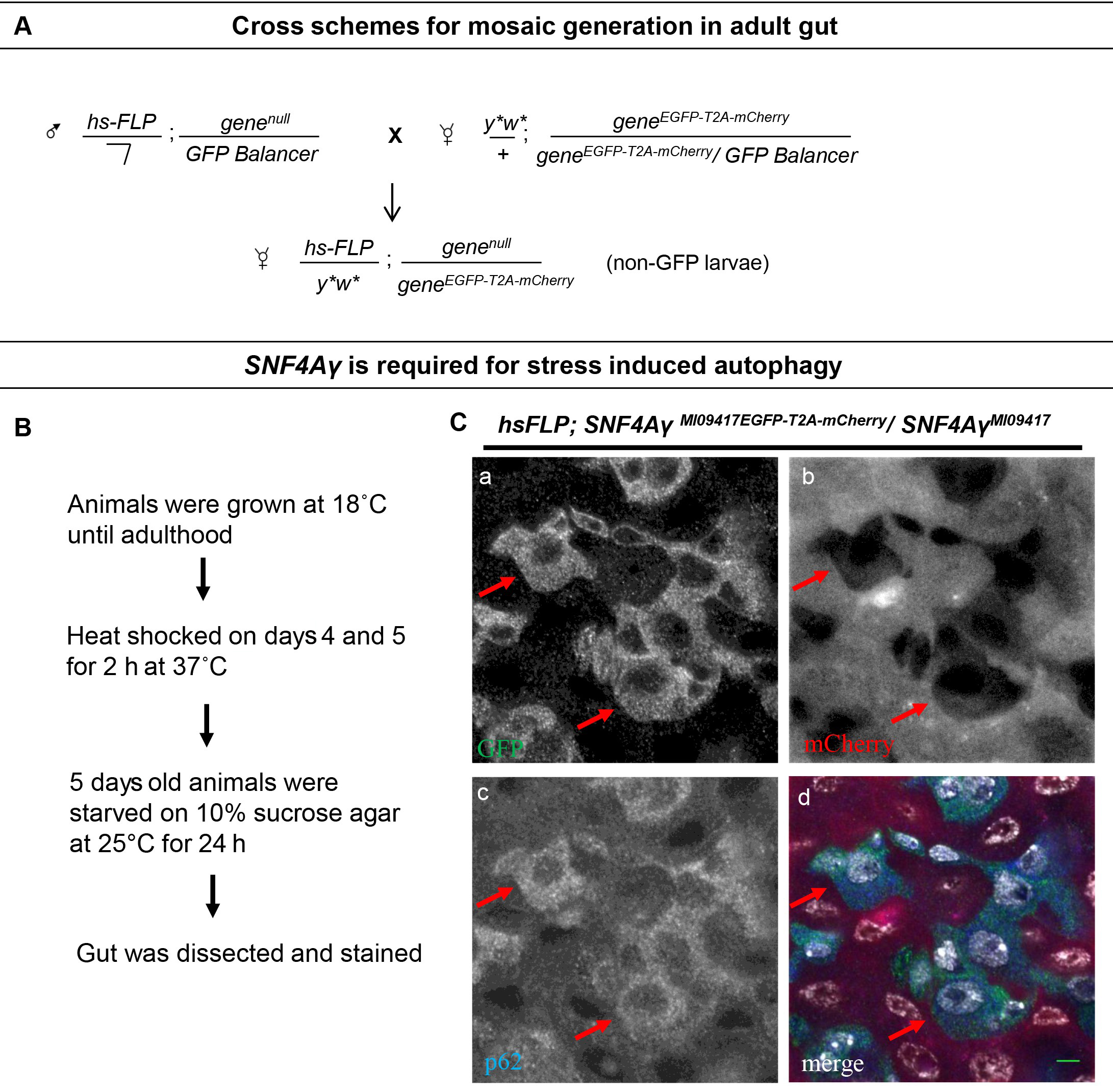
Figure 4. Random mosaic generation in adult gut. A. Schematic showing cross strategy for a random mosaic generation in adult gut. B. Experimental plan for starvation in adult flies. The image displaying the adult midgut from amino acid starved hsFLP;SNF4AγMI09417EGFP-T2A-mCherry/SNF4AγMI09417 animals, stained for GFP (a, d), mCherry (b, d), p62 (c, d) and DAPI (white) (d). Red arrows indicate control cells expressing SNF4Aγ-EGFP-SNF4Aγ, revealing higher levels of p62. In contrast, mutant cells are mCherry positive and have reduced p62 levels, indicating a defect in autophagy induction, scale bar = 5 µm. (Adapted from Nagarkar-Jaiswal et al., 2017)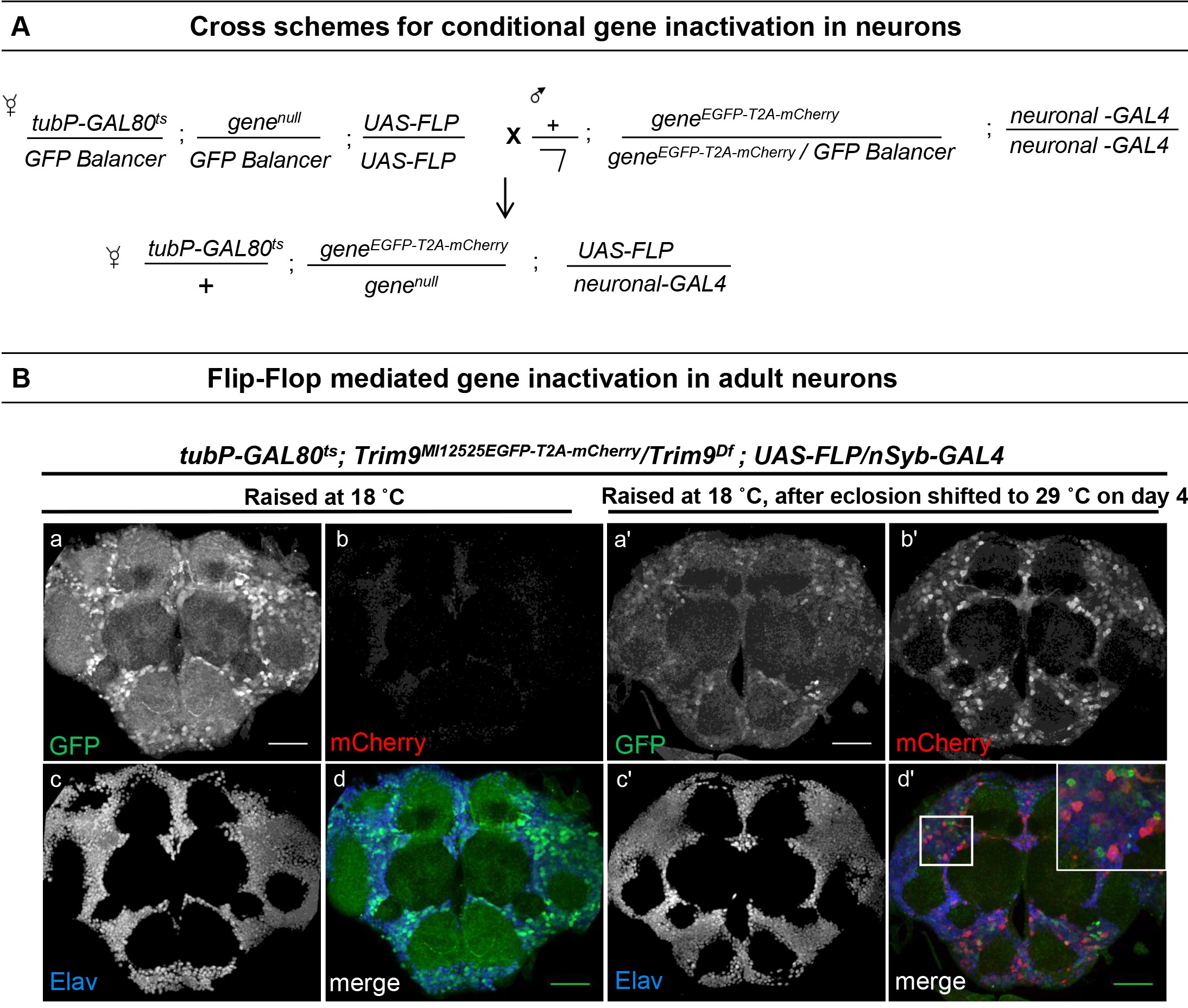
Figure 5. Gene inactivation in adult neurons. A. Crossing scheme for conditional gene inactivation in adult neurons combining the Flip-flop cassette with FLP expressed using the GAL4/GAL80/UAS binary system. B. Adult brain of tubP-GAL80ts;Trim9MI12525EGFP-T2A-mCherry/Trim9Df;UAS-FLP/nSyb-GAL4 animals stained for EGFP (green), mCherry (red) and Elav (blue). The temperature sensitive GAL80 (GAL80ts), expressed under a tubulin promoter (tubP) was used to prevent nSyb-GAL4 from driving expression of UAS-FLP during development. (a-d) Adult brains of tubP-GAL80ts;Trim9MI12525EGFP-T2A-mCherry/Trim9Df;UAS-FLP/nSyb-GAL4 control animals, raised at 18 °C display Trim9-EGFP-Trim9 (green, a and d) and do not show any mCherry expression (red, b and d). (a’-d’) Adult brains of tubP-GAL80ts;Trim9MI12525EGFP-T2A-mCherry/Trim9Df; UAS-FLP/nSyb-GAL4 animals, raised at 18 °C until four days after eclosion, and then shifted to 29 °C to induce FLP-expression, show mCherry-expressing neuronal cells in which the cassette inversion has taken place (red, b’ and d’), scale bars = 50 µm. (Adapted from Nagarkar-Jaiswal et al., 2017)
Recipes
- Grape juice plates
300 ml double distilled H2O
12 g agar
22 g sugar
100 ml Welch's grape juice concentrate
10 ml ethanol
5 ml glacial acetic acid- Add agar and sugar to cold ddH2O
- Microwave until dissolved (max. 12 min), stirring occasionally
- Add grape juice concentrate
- Cool to 65 °C and add Ethanol and acetic acid
- 10% sucrose agar plates
10 g sucrose
2 g agar
Make up the volume with water to 100 ml and microwave until dissolved, stirring occasionally
Acknowledgments
We thank Hugo J. Bellen for comments on the manuscript. SNJ and PP are supported by the Department of Science & Technology, Ramanujan Fellowship (SB/S2/RJN-157/2017). SNM is supported by NIH T32 (5T32HL098039-08).
Competing interests
The authors declare no competing interests.
References
- Kiehart, D. P., Crawford, J. M. and Montague, R. A. (2007). Collection, dechorionation, and preparation of Drosophila embryos for quantitative microinjection. CSH Protoc 2007: pdb prot4717.
- Kiehart, D. P., Crawford, J. M. and Montague, R. A. (2000). Quantitative microinjection of Drosophila embryos: general strategy. CSH Protoc 2000: pdb top5.
- Nagarkar-Jaiswal, S., Lee, P. T., Campbell, M. E., Chen, K., Anguiano-Zarate, S., Gutierrez, M. C., Busby, T., Lin, W. W., He, Y., Schulze, K. L., Booth, B. W., Evans-Holm, M., Venken, K. J., Levis, R. W., Spradling, A. C., Hoskins, R. A. and Bellen, H. J. (2015a). A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4: e05338.
- Nagarkar-Jaiswal, S., DeLuca, S. Z., Lee, P. T., Lin, W. W., Pan, H., Zuo, Z., Lv, J., Spradling, A. C. and Bellen, H. J. (2015b). A genetic toolkit for tagging intronic MiMIC containing genes. Elife 4: e08469.
- Nagarkar-Jaiswal, S., Manivannan, S. N., Zuo, Z. and Bellen, H. J. (2017). A cell cycle-independent, conditional gene inactivation strategy for differentially tagging wild-type and mutant cells. Elife 6: e26420.
- Venken, K. J., Schulze, K. L., Haelterman, N. A., Pan, H., He, Y., Evans-Holm, M., Carlson, J. W., Levis, R. W., Spradling, A. C., Hoskins, R. A. and Bellen, H. J. (2011). MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods 8(9): 737-743.
Article Information
Copyright
Manivannan et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Manivannan, S. N., Pandey, P. and Nagarkar-Jaiswal, S. (2019). Flip-flop Mediated Conditional Gene Inactivation in Drosophila. Bio-protocol 9(3): e3157. DOI: 10.21769/BioProtoc.3157.
- Nagarkar-Jaiswal, S., Manivannan, S. N., Zuo, Z. and Bellen, H. J. (2017). A cell cycle-independent, conditional gene inactivation strategy for differentially tagging wild-type and mutant cells. Elife 6: e26420.
Category
Molecular Biology > DNA > DNA modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










