- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of D-glutamate Production from the Dual Function Enzyme, 4-amino-4-deoxychorismate Lyase/D-amino Acid Transaminase, in Mycobacterium smegmatis
Published: Vol 9, Iss 1, Jan 5, 2019 DOI: 10.21769/BioProtoc.3135 Views: 5799
Reviewed by: Valentine V TrotterWilliam Ryan WillAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views

Activation of X-Succinate Synthases for Fumarate Hydroalkylation Using an In Vitro Activation Method
Anshika Vats [...] Mary C. Andorfer
Jun 20, 2025 2510 Views

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2441 Views
Abstract
D-amino acid transaminase (D-AAT) is able to synthesize both D-glutamate and D-alanine, according to the following reaction: D-alanine + α-ketoglutarate ⇌ D-glutamate + pyruvate. These two D-amino acids are essential components of the peptidoglycan layer of bacteria. In our recently published work, MSMEG_5795 from Mycobacterium smegmatis was identified as having D-amino acid transaminase (D-AAT) activity, although it has primarily been annotated as 4-amino-4-deoxychorismate lyase (ADCL). To unequivocally demonstrate D-AAT activity from MSMEG_5795 protein two coupled enzyme assays were performed in series. First, D-alanine and α-ketoglutarate were converted to D-glutamate and pyruvate by MSMEG_5795 using the D-AAT assay. Next, the products of this reaction, following removal of all protein, were used as input into an assay for glutamate racemase in which D-glutamate is converted to L-glutamate by glutamate racemase (Gallo and Knowles, 1993; Poen et al., 2016). As the only source of D-glutamate in this assay would be from the reaction of D-alanine with MSMEG_5795, positive results from this assay would confirm the D-AAT activity of MSMEG_5795 and of any enzyme tested in this manner.
Keywords: D-amino acid transaminaseBackground
The protocol described here in detail was developed during a study of suppressor mutants in M. smegmatis strain in which glutamate racemase (murI) had been deleted (Mortuza et al., 2018). During this work, it became apparent that a mutation in the promoter of 4-amino-4-deoxychorismate lyase (ADCL) (MSMEG_5795) was unexpectedly able to complement the murI deletion. This mutation resulted in a more than 10-fold increased expression of MSMEG_5795. Because ADCL is homologous to D-amino acid transaminase (D-AAT), we decided to test MSMEG_5795 for D-AAT activity. The initial test which involved incubation of the enzyme with D-alanine and α-ketoglutarate was positive for activity. To be doubly sure that this protein, which was predominantly annotated as ADCL, was actually producing glutamate, we used the output of the D-AAT assay as input to the previously reported coupled assay for MurI (Tanizawa et al., 1987; Gallo and Knowles, 1993). This assay is detailed below.
Materials and Reagents
- Pipette tips (Labcon, 1,250 µl, catalog number: 1045-260-300; 200 µl, catalog number: 1030-260-300; 10 µl, catalog number: 1036-260-000)
- Microcentrifuge tubes 1.7 ml (MultiMax, catalog number: 2942)
- 50 ml conical centrifuge tubes (Cellstar, catalog number: 227261)
- Cuvette Type 9 quartz 10 mm path (Starna, catalog number: 9Q10)
- VivaspinTM 2 protein concentrator spin column, MWCO 10,000 Da (GE Healthcare Life Sciences, catalog number: 28932247)
- Syringe filter, 0.45 µm (Ahlstrom, catalog number: 760506)
- Syringe with Luer-Lok tip, 20 ml (Becton Dickinson, catalog number: 3028360)
- 10-15 mg/ml of purified Mycobacterium smegmatis MSMEG_5795 in 20 mM Tris-HCl pH 8.0 and 150 mM NaCl (Mortuza et al., 2018), and 20-40 mg/ml of purified Bacillus anthracis glutamate racemase isoform 2 (MurI2Ba) protein in 50 mM Tris pH 8.0, 250 mM NaCl and 0.1 mM DTT (May et al., 2007)
- Tris base (NeoFroxx, catalog number: 1125KG001)
- HCl, 37%, reagent grade, ACS (Scharlau, catalog number: AC07412500)
- α-ketoglutaric acid sodium salt (Sigma-Aldrich, catalog number: K1875)
- Pyridoxal 5′-phosphate hydrate (PLP) (Sigma-Aldrich, catalog number: P9255)
- L-lactate dehydrogenase from hog muscle (Roche, catalog number: 10107085001)
- β-Nicotinamide adenine dinucleotide, reduced dipotassium salt (NADH) (Sigma-Aldrich, catalog number: N4505)
- D-alanine (Sigma-Aldrich, catalog number: A7377)
- NaOH (ACS reagent grade, catalog number: SO042500)
- N-Cyclohexyl-2-aminoethanesulfonic acid (CHES) (ACROS, catalog number: 208181000)
- Iodonitrotetrazolium chloride (INT) (Sigma-Aldrich, catalog number: I10406)
- Diaphorase (Sigma-Aldrich, catalog number: D5540)
- Nicotinamide adenine dinucleotide (NAD+) (Sigma-Aldrich, catalog number: N1511)
- Adenosine 5’-diphosphate (ADP) (Sigma-Aldrich, catalog number: A2754)
- L-glutamate dehydrogenase (LGDH) (Sigma-Aldrich, catalog number: G2501)
- PierceTM Coomassie Plus (Bradford) Assay Kit (Thermo Fisher, catalog number: 23236)
- 10 M NaOH (made by gradually dissolving NaOH pellets in ASTM Type I (18 MΩ) water, stored at room temperature, no need to sterilize)
Note: All solutions below are prepared in ASTM Type I (18 MΩ) water and filtered through 0.45 μm filters prior to use. Solutions are stored long-term at 4 °C, unless otherwise noted.
- 1 M Tris base (adjust the solution to pH 8.1 with HCl)
- 0.5 M α-ketoglutaric acid
- 2 mM PLP
- 10 mM NADH (prepare this solution fresh for each assay)
- 0.5 M D-alanine
- 1 M CHES (adjust the solution to pH 9.2 with NaOH)
- D-amino acid transaminase reaction partial mixture (see Recipes)
- Glutamate reaction partial mixture (see Recipes)
Equipment
- Magnetic stir bars 15 mm x 6 mm (BRAND, catalog number: 137114)
- Pipetman Neo® Pipets (Gilson, model: P1000N, catalog number: F144566; P200N, catalog number: F144565; P20N, catalog number: F144563)
- 25 ml glass beaker (Boeco, catalog number: BOE 5010614)
- Microcentrifuge (Eppendorf, model: 5415R)
- Magnetic stirrer (Barnstead Thermolyne, model: SP131010-33)
- UltrospecTM 3100 pro UV/Visible Spectrophotometer with SWIFT II software (GE Healthcare Life Sciences, model: UltrospecTM 3100 pro)
- MultiTemp III (GE Healthcare) thermostatic circulator to control the temperature of the UV-Vis spectrophotometer cuvettes
- -20 °C freezer
Software
- BiochromTM SWIFT II Software, version 2.05
Procedure
- Assay Preparations
- Purify 10-15 mg/ml MSMEG_5795 and 20-40 mg/ml MurI2Ba protein using immobilized metal affinity chromatography and size exclusion chromatography.
Note: Determine the mycobacterial protein concentration using the PierceTM Coomassie Plus (Bradford) Assay according to the manufacturer’s recommendation. Centrifuge both proteins at 16,000 x g for 20 min at 4 °C to remove any aggregates prior to being assayed. - Prepare all stock solutions for the assays (see Recipes).
- Purify 10-15 mg/ml MSMEG_5795 and 20-40 mg/ml MurI2Ba protein using immobilized metal affinity chromatography and size exclusion chromatography.
- D-amino acid transaminase reaction
- Prepare desired volume of the D-AAT assay reaction partial mixture containing 100 mM Tris-HCl pH 8.1, 10 mM α-ketoglutarate, 0.15 mM PLP, 5 U L-lactate dehydrogenase, 0.2 mM NADH and 77 nM MSMEG_5795 protein. For example, 20 ml is a convenient amount to make here. As noted above the NADH is prepared fresh each day before conducting the assay.
Note: Keep assay mix on ice during assay and equilibrate to room temperature before use. - Transfer 950 µl of the reaction mixture to a 1 ml quartz cuvette and pre-incubate at 30 °C for 5 min.
- Measure the absorbance at 340 nm for 2 min to set a baseline on the UltrospecTM 3100 pro.
- To start the reaction, add 50 µl 0.5 M D-alanine substrate to the assay mix to a final concentration of 25 mM to start the reaction and incubate each cuvette for 20, 40 and 60 min at 30 °C. The absorbance should be decreasing with time.
- Add 50 µl water instead of substrate to the negative control sample and incubate for 60 min at 30 °C.
- Following incubation, transfer the cuvettes immediately to 4 °C to halt the reaction.
- Filter the reaction mix using a Vivaspin 2 protein concentrator spin column with MWCO of 10,000 Da to remove MSMEG_5795 and L-lactate dehydrogenase. Use this protein-free output as a substrate for the subsequent glutamate racemase (MurI) activity assay.
Note: Samples can be stored at -20 °C before second coupled enzyme assay is performed or for further analysis.
- Prepare desired volume of the D-AAT assay reaction partial mixture containing 100 mM Tris-HCl pH 8.1, 10 mM α-ketoglutarate, 0.15 mM PLP, 5 U L-lactate dehydrogenase, 0.2 mM NADH and 77 nM MSMEG_5795 protein. For example, 20 ml is a convenient amount to make here. As noted above the NADH is prepared fresh each day before conducting the assay.
- Glutamate racemase reaction
- Prepare desired volume of MurI assay reaction partial mixture containing 50 mM CHES pH 9.2, 5 mM NAD+, 37.5 U ml-1 L-glutamate dehydrogenase, 2.5 mM ADP, 0.65 mM INT, 2 U ml-1 diaphorase. For example, 10 ml is a convenient amount to make here.
Note: Mix assay components vigorously in a glass beaker, but avoid foaming by using a small stir bar and magnetic stirrer. Filter mixture with a 0.45 µm syringe filter and a 20 ml syringe with a Luer-Lok tip. Keep solution on ice during assay. Equilibrate to room temperature before use. This solution will take on a light salmon hue. - For each reaction sample, transfer 875 µl of the reaction mixture to a 1 ml quartz cuvette.
- Add 100 µl of the product coming from the D-AAT assay to each cuvette.
- Start the reaction by adding 25 µl MurI2Ba to a final enzyme concentration of 10 µM.
- To the blank sample, add the buffer used for the MurI2Ba stock solution in place of MurI2Ba and add water instead of substrate.
- Add water instead of substrate to the negative control sample.
- Monitor the absorbance at 500 nm for 30 min at 30 °C.
Note: Subtract the absorbance values obtained from the blank sample from each measurement to obtain the final absorbance values.
- Prepare desired volume of MurI assay reaction partial mixture containing 50 mM CHES pH 9.2, 5 mM NAD+, 37.5 U ml-1 L-glutamate dehydrogenase, 2.5 mM ADP, 0.65 mM INT, 2 U ml-1 diaphorase. For example, 10 ml is a convenient amount to make here.
Data analysis
The product of the D-AAT assay is taken as input for the glutamate racemase assay to confirm the presence of glutamate by monitoring the absorbance increase at 500 nm. The increase in absorbance values for the 20, 40 and 60 min samples corresponds to the expected increase in glutamate production over time (Figure 1). The detection of glutamate as reported here is qualitative. This result was also supported with data obtained from high-resolution mass spectrometry of the assay output which showed an increase in glutamate concentration of 0.3, 0.7 and 1.0 mM in samples incubated for 20, 40 and 60 minutes respectively. As a result, the ability to convert D-alanine to D-glutamate by the MSEMG_5795 enzyme can be assessed using the glutamate racemase assay. 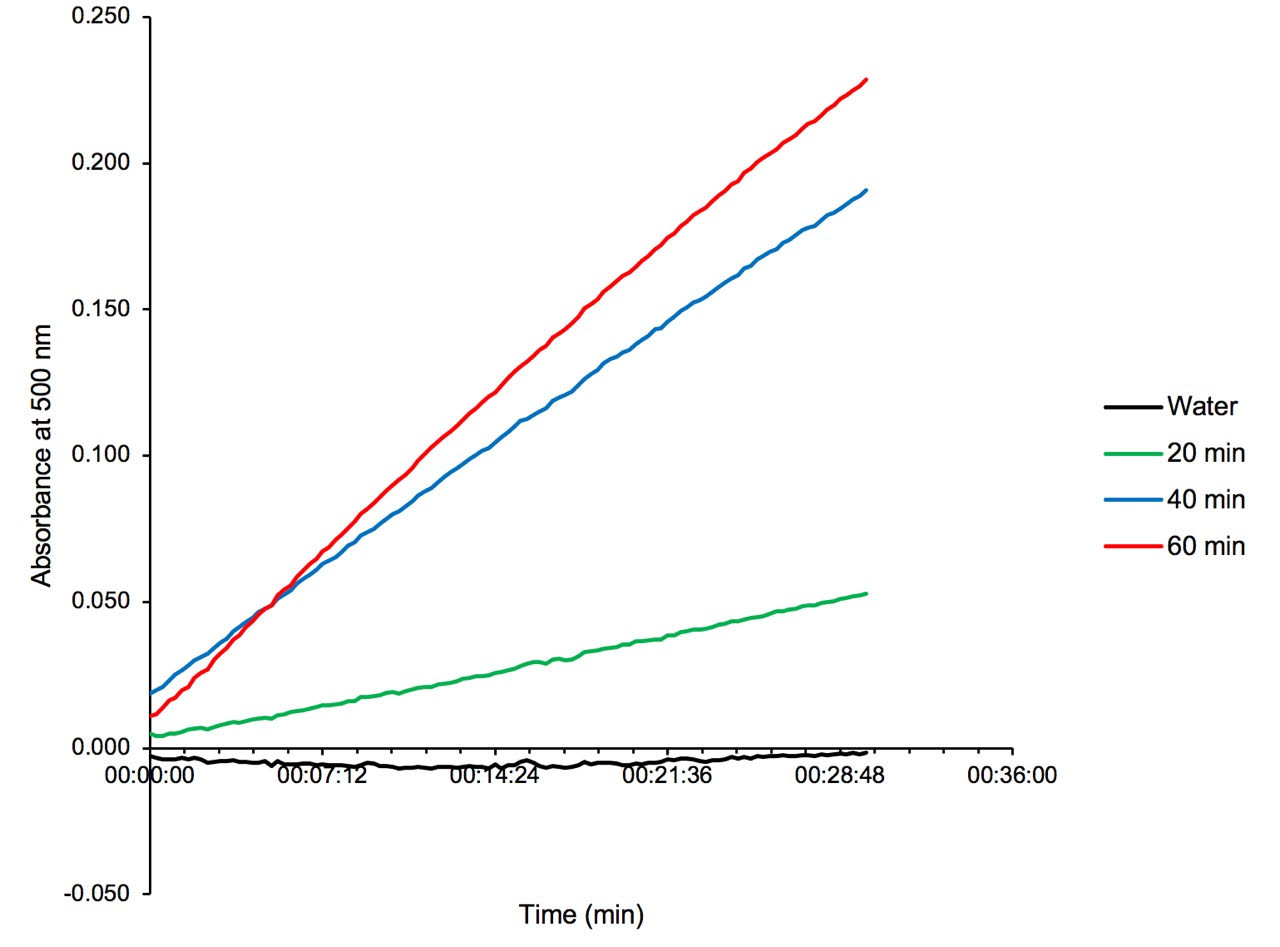
Figure 1. Time-dependent glutamate production by MSMEG_5795 verified with glutamate racemase enzyme assay (see Step C7). Protein-free product from the D-AAT assay was used as substrate for the glutamate racemase assay in the D to L direction. The increase in absorbance was observed at 500 nm for 30 min. Each sample was taken in duplicate and contained 100 µl substrate and 10 µM glutamate racemase. Blank sample was subtracted from each measurement and absorbance values were averaged before plotting against time. Absorbance values obtained from the 20, 40 and 60 min samples are shown in green, blue and red respectively. The control sample containing water instead of substrate is shown in black. Figure 1 is originally published in Mortuza et al., 2018. (This figure is reprinted with permission from Molecular Microbiology.)
Notes
- The concentrations of the components listed in the assay mixtures represent the desired concentrations to have in place at the start of the assay, but be sure to take into account any dilution that may result from the addition of enzyme or other substance added subsequently.
- If there is any concern that L-glutamate might be produced by the first enzyme being tested in this assay an additional control can be added in which the output of the first enzyme is assayed using the enzyme L-glutamate dehydrogenase directly without incubation with MurI.
- Other glutamate racemases that are sufficiently active at 30 °C and pH 9.2 would likely work here as well, but we have only trialed this assay with the MurI2Ba enzyme.
Recipes
- D-amino acid transaminase reaction partial mixture
100 mM Tris-HCl, pH 8.1
10 mM α-ketoglutarate
0.15 mM PLP
5 U L-lactate dehydrogenase
0.2 mM NADH
77 nM MSMEG_5795 protein
Prepare in ASTM Type I (18 MΩ) water
Filter through a 0.45 μm filter prior to use
Long-term storage not recommended. Prepare fresh each day
Substrate is added later - Glutamate reaction partial mixture
50 mM CHES, pH 9.2
5 mM NAD+
37.5 U ml-1 L-glutamate dehydrogenase
2.5 mM ADP
0.65 mM iodonitrotetrazolium chloride
2 U ml-1 diaphorase
Prepare in ASTM Type I (18 MΩ) water
Filter through a 0.45 μm filter prior to use
Long-term storage not recommended. Prepare fresh each day
Substrate and MurI are added later
Acknowledgments
This protocol was first described in brief in Mortuza et al. (2018). This research was supported by Lottery Health Research New Zealand-LHR2017-48905, the University of Otago, the Maurice Wilkins Centre-University of Auckland and the Thrash Foundation, Houston, Texas.
Competing interests
The authors have no conflicts to declare.
References
- Gallo, K. A. and Knowles, J. R. (1993). Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry 32(15): 3981-3990.
- May, M., Mehboob, S., Mulhearn, D. C., Wang, Z., Yu, H., Thatcher, G. R., Santarsiero, B. D., Johnson, M. E. and Mesecar, A. D. (2007). Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design. J Mol Biol 371(5): 1219-1237.
- Mortuza, R., Aung, H. L., Taiaroa, G., Opel-Reading, H. K., Kleffmann, T., Cook, G. M. and Krause, K. L. (2018). Overexpression of a newly identified D-amino acid transaminase in Mycobacterium smegmatis complements glutamate racemase deletion. Mol Microbiol 107(2): 198-213.
- Poen, S., Nakatani, Y., Opel-Reading, H. K., Lasse, M., Dobson, R. C. and Krause, K. L. (2016). Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery. Biochem J 473(9): 1267-1280.
- Tanizawa, K., Masu, Y., Asano, S., Tanaka, H. and Soda, K. (1987). D-Amino acid aminotransferase from a thermophile, YM-1: enzymological properties, cloning of the amino acid sequence. In: The 7th International Congress on Chemical and Biological Aspects of Vitamin B6 Catalysis. In: Korpela, T. and Christen, P. (Eds). Turku, Finland Birkhauser Verlag: 43-46.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Opel-Reading, H. K., Mortuza, R. and Krause, K. L. (2019). Detection of D-glutamate Production from the Dual Function Enzyme, 4-amino-4-deoxychorismate Lyase/D-amino Acid Transaminase, in Mycobacterium smegmatis. Bio-protocol 9(1): e3135. DOI: 10.21769/BioProtoc.3135.
Category
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Other compound > Amino acid
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









