- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Using Arabidopsis Mesophyll Protoplasts to Study Unfolded Protein Response Signaling
Published: Vol 8, Iss 23, Dec 5, 2018 DOI: 10.21769/BioProtoc.3101 Views: 6420
Reviewed by: Amey RedkarHua LiuPengxiang Fan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification of S-locus F-box Protein Sequences in Diploid Potato, Solanum okadae, via Degenerate PCR
Amar Hundare [...] Timothy P. Robbins
Jun 5, 2025 1995 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2020 Views

Isolation and Transfection of Protoplasts From Maize Mesophyll Cells
Lauren A. Higa [...] Zhi-Yan Du
Feb 5, 2026 64 Views
Abstract
Various environmental stresses or artificial reagents can trigger unfolded protein accumulation in the endoplasmic reticulum (ER) due to the folding capacity of the ER being exceeded. This is termed ER stress, and triggers the unfolded protein response (UPR). Assays for activation of the UPR in plants include Tunicamycin (Tm)- or dithiothreitol (DTT)-mediated root growth inhibition, analysis of splicing of the UPR-responsive transcription factor bZIP60 (basic Leucine Zipper Domain 60), and upregulation of relevant UPR genes. We provide here a quick and robust method to detect UPR signaling in Arabidopsis thaliana protoplasts. This assay can also be applied to other plant species for which protoplasts can be isolated.
Keywords: ArabidopsisBackground
Substantial progress has been made over the last two decades in our understanding of the UPR in plants. In addition to its important role in stress responses (Howell, 2013; Bao et al., 2017; Bao and Howell, 2017), the UPR also participates in plant reproductive development, vegetative growth and root development (Deng et al., 2013; Deng et al., 2016; Kim et al., 2018). There are two arms to the UPR in plants that employ three transcription factors: bZIP60, which is activated by IRE1 (Inositol Requiring 1) -mediated splicing of its mRNA (Deng et al., 2011), and bZIP17 and bZIP28, which are activated by S1P- and S2P-mediated cleavage of their membrane-localized proteins (Liu et al., 2007; Srivastava et al., 2013). All three transcription factors induce expression of downstream genes, including molecular chaperones such as BIP3 that assist in protein folding.
Protoplasts can be easily isolated from various plant species, allowing transient expression in protoplasts of potential factors involved in UPR signaling, in place of the time-consuming generation of transgenic plants. In our recent paper on ER stress-regulated autophagy mediated by IRE1b, we successfully detected bZIP60 splicing and BIP3 expression in Arabidopsis protoplasts (Bao et al., 2018) as hallmarks of the UPR (Iwata and Koizumi, 2005; Iwata et al., 2008; Deng et al., 2011), and used this assay to determine the effect of expression of IRE1b mutants. Here, we describe this assay in detail; we believe the robustness and reproducibility of this assay will facilitate plant UPR research.
Materials and Reagents
- Aluminum Foil (Reynolds Kitchens, REYNOLDS WRAP, catalog number: 353224)
- 6-well Plate (Corning, Falcon, catalog number: 353224)
- 12-well Plate (Corning, Falcon, catalog number: 353225)
- 100 x 15 mm Square Petri Dish (Corning, Falcon, catalog number: 351112)
- 14 ml Polypropylene Round-Bottom Tube (Corning, Falcon, catalog number: 352059)
- Electrical tape (ULINE, Hyper Tough, catalog number: 34366)
- Rosette leaves of wild-type Arabidopsis thaliana (WT, Columbia-0 ecotype)
- ire1a ire1b mutant Arabidopsis plants
- IRE1b plasmids
- bZIP60s, BIP3 and Actin2 specific primers
Primers for detecting bZIP60s (spliced form of bZIP60: bZIP60F4 and bZIP60SB2), BIP3 and Actin2:
bZIP60F4: GAAGGAGACGATGATGCTGTGGCT
bZIPSB2: AGCAGGGAACCCAACAGCAGACT
BIP3F: TTCGACCCGAAACGTCTGATTGGA
BIP3R: GCTTGCCTCTGCGCATCATTGAAA
Actin2F: GGAAGGATCTGTACGGTAAC
Actin2R: GGACCTGCCTCATCATACT - Maxiprep Kit (Gene Elute HP Plasmid) (Sigma-Aldrich, catalog number: NA310-1KT)
- RNA Extraction Kit (DNeasy Plant Mini Kit) (QIAGEN, catalog number: 69104)
- cDNA kit (iScriptTM cDNA Synthesis Kit) (Bio-Rad, catalog number: 1708890)
- Tunicamycin (Tm) (Sigma-Aldrich, catalog number: T7765)
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 3483-12-3)
- PEG4000 (PEG) (Sigma-Aldrich, catalog number: 25322-68-3)
- Cellulose R-10 (Yakult, CELLULASE “ONOZUKAR-10”)
- Macerozyme R-10 (Yakult, MACEROZYME R-10)
- BSA (Sigma-Aldrich, Bovine Serum Albumin, catalog number: 9048-46-8)
- DMSO (Sigma-Aldrich, dimethyl sulfoxide, catalog number: 67-68-5)
- Mannitol (Sigma-Aldrich, catalog number: 69-65-8)
- β-mercaptoethanol (Sigma-Aldrich, catalog number: 60-24-2)
- MgCl2 (Sigma-Aldrich, Magnesium chloride, catalog number: 7791-18-6)
- MES (Sigma-Aldrich, catalog number: 4432-31-9)
- NaCl (Sigma-Aldrich, Sodium chloride, catalog number: 7647-14-5)
- CaCl2 (Sigma-Aldrich, Calcium chloride, catalog number: 10043-52-4)
- KCl (Sigma-Aldrich, Potassium chloride, catalog number: 7447-40-7)
- KOH (Sigma-Aldrich, Potassium hydroxide, catalog number: 1310-58-3)
- 2x YT media (see Recipes)
- Yeast Extract (BD Biosciences, BD Bacto, catalog number: 212750)
- Tryptone (Neogen Food Safety, Acumedia, catalog number: 7351A)
- Enzyme solution (see Recipes)
- MMg buffer (see Recipes)
- 40% PEG (see Recipes)
- W5 solution (see Recipes)
- Stock solutions (see Recipes)
Equipment
- Pipette (Eppendorf, Eppendorf Research Plus, catalog number: 2231000222)
- Incubator (New Bruswick Scientific, Innova42, catalog number: M1335-0000)
- Hemocytometer (Hausserscientific, Bright-Line Counting Chambers, catalog number: 3100)
- Microscope (Nikon, Eclipse TS100)
- High Speed Centrifuge (Thermo Scientific, model: SorvallTM RC 6 Plus, catalog number: 36-101-0816)
- Low Speed Centrifuge (Thermo Scientific, model: HereusTM MultifugeTM X3R, catalog number: 75004516)
- Microcentrifuge (Thermo Scientific, model: SorvallTM LegendTM Micro 17, catalog number: 75002403)
- NanoDrop Spectrophotometer (Scientific Industries, model: Vortex-Genie 2, catalog number: SI-0236)
- Thermocycler (S1000 Thermal Cycler) (Bio-Rad, catalog number: 1852196)
- DNA Gel electrophoresis cell (Bio-Rad, Sub Cell GT Cell, catalog number: 1640302, or similar)
- DNA Gel scanner (FOTODYNE, FOTO/Analyst ImageTech, catalog number: 6-9030S2DL, or similar)
Software
- ImageJ
- Adobe Photoshop
Procedure
- Prepare plasmids for expression of genes of interest (IRE1b is used as an example in this protocol). For each extraction, grow E. coli in 500 ml 2x YT media overnight and extract using a maxiprep kit (QIAGEN product) according to the manufacture's protocol. Set the DNA concentration to 1 μg/μl.
- All buffers and chemicals should be prepared at least one day before protoplast transformation (Yoo et al., 2007).
- Make fresh enzyme solution with Cellulose R-10 and Macerozyme R-10 (Products of Yakult).
- Prepare protoplasts from rosette leaves of wild-type (WT) and/or mutant Arabidopsis plants (Yoo et al., 2007) (ire1a ire1b is used as an example in this protocol) using the tape method as described by Wu et al. (2009) (Figures 1A-1C).
- Load 10 μl of cell suspension in a hemocytometer and count cells under a light microscope; resuspend protoplasts in MMg buffer to a density of 2.5 x 106/ml.
- For each reaction, in a 2 ml round bottom tube, mix 400 μl protoplasts with 30 μl IRE1b plasmid (1 μg/μl), add an equal volume of 40% PEG (430 μl, see Recipes for making 40% PEG), mix and incubate at room temperature for 10 min (Figure 1D). Spin down at 60 x g and wash twice with W5 to remove PEG.
- Resuspend with 1 ml W5 and transfer the transformed protoplasts into a 6-well culture plate (or 12-well culture plate if more than 6 samples), add W5 to a final volume of 5 ml (Figure 1E), and incubate overnight in the dark by covering with foil.
- Carefully discard the supernatant by gentle pipetting (protoplasts will stay on the bottom due to gravity, no need to centrifuge), and re-suspend with W5 to a final volume of 3 ml.
- Divide the 3 ml protoplasts equally into two 2 ml round bottom tubes; add Tm to 5 μg/ml or DTT to 2 mM, mix the protoplasts by gentle inversion, and incubate for 6 h, mixing every 2 h.
- Spin at 60 x g for 1 min in a low speed centrifuge, discard the supernatant.
- Add RNA extraction buffer (QIAGEN, Plant Mini RNA kit) and vortex thoroughly for 30 s. Extract RNA according to the manufacturer’s protocol.
- Test the concentration and quality of RNA using a NanoDrop Spectrophotomer; if desired, RNA can be stored at -80 °C for later use.
- Use 0.5 μg RNA to synthesize cDNA according to the manufacturer’s protocol (Bio-Rad cDNA kit).
- Detect the bZIP60 spliced form (named bZIP60s) by RT-PCR using bZIP60F4 and bZIP60SB2 primers, and BIP3 expression using BIP3F and BIP3 primers. Actin2 (using Actin2F and Actin2R primers) is used as a loading control.
- Run the PCR products on 1.5% agarose gel, stain with Ethidium Bromide and visualize using a gel scanner under UV light (Figure 1F). qRT-PCR can be used for quantification if needed, normalized against Actin2.
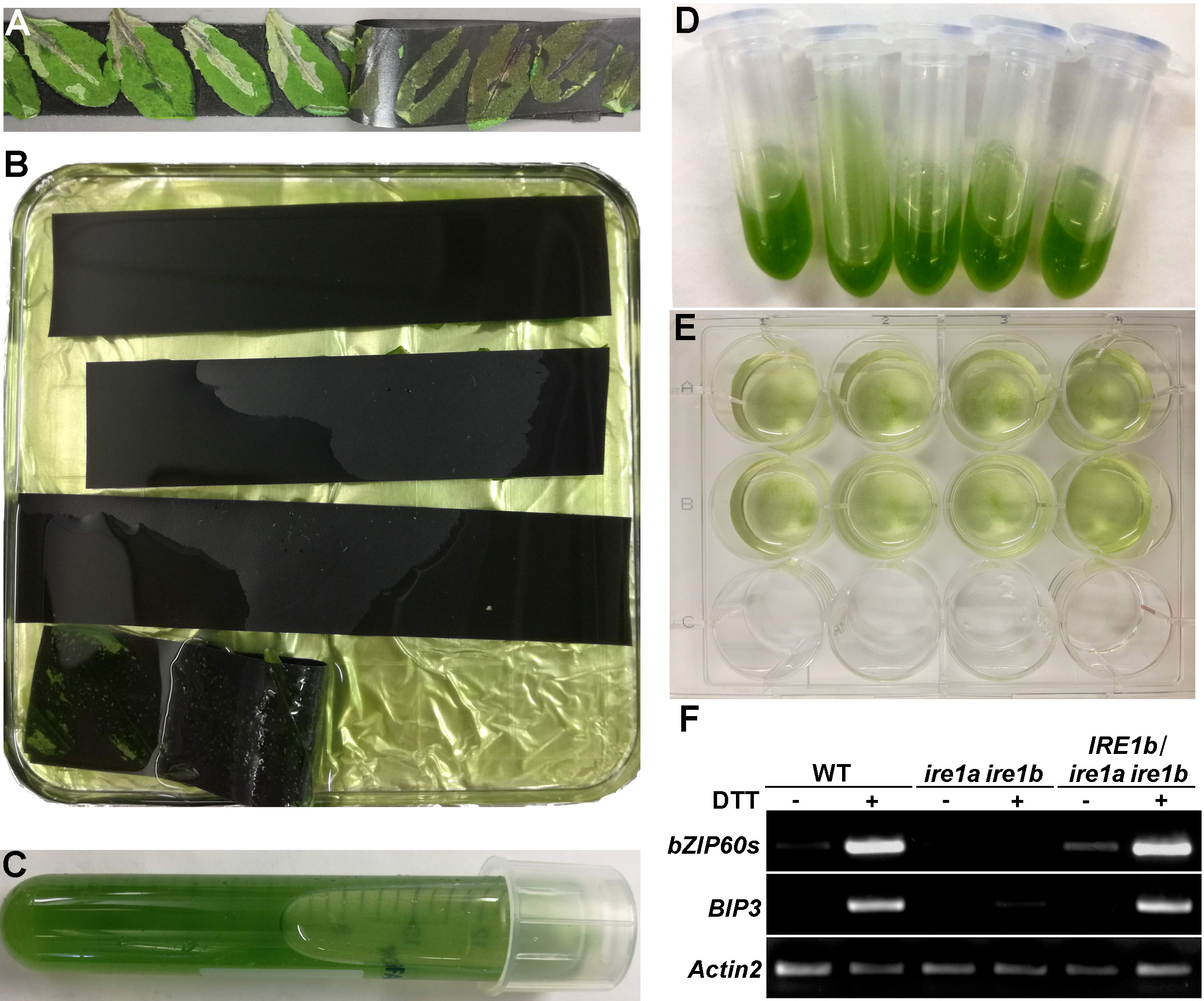
Figure 1. An overview of Arabidopsis protoplast isolation and transformation with the resulting RT-PCR. A-B. Taped rosette leaves before (A) and after (B) digestion with enzymes; C. Isolated protoplasts in W5 solution; D. Protoplasts during transformation; E. Transformed protoplasts after transfer to a 12-well plate; F. RT-PCR detection of bZIP60s and BIP3 expression in protoplast samples treated with or without 2 mM DTT for 6 h. IRE1b was introduced into an ire1a ire1b double mutant background and showed restoration of bZIP60 splicing and BIP3 induction. Actin2 was employed as an endogenous loading control.
Data analysis
Three independent transformations are required for each experiment, images are taken and quantified using ImageJ (Reference 15) if needed, and significant differences assessed by Student’s t-test.
Notes
- Use round bottom tubes (size: 2 ml and 14 ml) for all experiments.
- To make sure the protoplast suspension is equally divided into two, pipette slowly and consistently.
- The product of the spliced form of bZIP60 (bZIP60s) in this assay is small; a 1.5% gel or higher is recommended.
- If RNA quantities are low, combine two transformations for RNA extraction for each construct.
- The use of an ire1a ire1b double mutant as a control is highly recommended, as splicing of bZIP60 is completely blocked in this mutant background.
- For isolation of protoplasts from rosette leaves, four-week-old healthy and well-expanded leaves from plants prior to flowering are preferable.
- PEG/Ca2+ solution is used to permeabilize the protoplast membrane to allow DNA uptake.
- Place transformed protoplasts in the dark to minimize potential light stress.
Recipes
- 2x YT media
16 g/L Tryptone
10 g/L yeast extract
5 g/L NaCl - Enzyme solution
- Make enzyme solution fresh with a final concentration of 0.4 M Mannitol, 20 mM KCl, 20 mM MES, 1.5% (w/v) cellulase R-10, 0.3% (w/v) macerozyme R-10
- Heat the enzyme solution in a 55 °C water bath for 10 min, then cool to room temperature before adding 10 mM CaCl2, 5 mM β-mercaptoethanol and 0.1% BSA (w/v)
- MMg buffer
400 mM mannitol
15 mM MgCl2
4 mM MES (Adjust pH to 5.7 with KOH) - 40% PEG
40% PEG4000 (w/v)
200 mM mannitol
100 mM CaCl2 - W5 solution
154 mM NaCl
125 mM CaCl2
5 mM KCl
2 mM MES
Adjust pH to 5.7 with KOH - Stock solutions
- Make 10 mg/ml Tm (in DMSO) and 1 M DTT (in ddH2O) stock solutions and store at -20 °C. Do NOT refreeze DTT more than 3 times
- Make 1 M mannitol, 0.3 M MgCl2, 1 M CaCl2, 0.5 M MES (Adjust pH to 5.7 with KOH), 1 M NaCl, 1 M KCl stock solutions and keep at room temperature
Acknowledgments
This work was supported by the National Science Foundation under Grant IOS 1353867 to DCB.
Competing interests
Authors claim no conflict of interest in this protocol.
References
- Bao, Y. and Howell, S. H. (2017). The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front Plant Sci 8(344) DOI: 10.3389/fpls.2017.00344.
- Bao, Y., Mugume, Y. and Bassham, D. C. (2017). Biochemical methods to monitor autophagic responses in plants. Methods Enzymol 588: 497-513.
- Bao, Y., Pu, Y., Yu, X., Gregory, B. D., Srivastava, R., Howell, S. H. and Bassham, D. C. (2018). IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy: 1-12.
- Deng, Y., Humbert, S., Liu, J., Srivastava, R., Rothstein, S. and Howell, S. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. PNAS 108(17): 7247-7252.
- Deng, Y., Srivastava, R. and Howell, S. (2013). Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. PNAS 110(48): 19633-19638.
- Deng, Y., Srivastava, R., Quilichini, T. D., Dong, H., Bao, Y., Horner, H. T. and Howell, S. H. (2016). IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J 88(2): 193-204.
- Howell, S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477-499.
- Iwata, Y., Fedoroff, N. V. and Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20(11): 3107-3121.
- Iwata, Y. and Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. PNAS 102(14): 5280-5285.
- Kim, J. S., Yamaguchi-Shinozaki, K. and Shinozaki, K. (2018). ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol 176(3): 2221-2230.
- Liu, J. X., Srivastava, R., Che, P. and Howell, S. H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19(12): 4111-4119.
- Srivastava, R., Deng, Y., Shah, S., Rao, A. G. and Howell, S. H. (2013). BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25(4): 1416-1429.
- Wu, F. H., Shen, S. C., Lee, L. Y., Lee, S. H., Chan, M. T. and Lin, C. S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16.
- Yoo, S. D., Cho, Y. H. and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7): 1565-1572.
- Rasband, W.S. ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/, 1997-2018.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bao, Y. and Bassham, D. C. (2018). Using Arabidopsis Mesophyll Protoplasts to Study Unfolded Protein Response Signaling. Bio-protocol 8(23): e3101. DOI: 10.21769/BioProtoc.3101.
Category
Plant Science > Plant molecular biology > Protein
Plant Science > Plant cell biology > Cell isolation
Cell Biology > Cell signaling > Stress response
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link