- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Papain-based Single Cell Isolation of Primary Murine Brain Endothelial Cells Using Flow Cytometry
Published: Vol 8, Iss 22, Nov 20, 2018 DOI: 10.21769/BioProtoc.3091 Views: 11881
Reviewed by: Pengpeng LiRAVI THAKURPatrick Ovando-Roche

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2361 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 1984 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2233 Views
Abstract
Brain endothelial cells (BECs) form the integral component of the blood-brain barrier (BBB) which separates the systemic milieu from the brain parenchyma and protects the brain from pathogens and circulating factors. In order to study BEC biology, it was of particular interest to establish a method that enables researchers to investigate and understand the underlying molecular mechanisms regulating their function during homeostasis, aging and disease. Furthermore, due to the heterogeneity of the cerebrovasculature and different vessel types that comprise the BBB, it is of particular interest to isolate primary BECs for single cell analysis from various subregions of the brain, such as the neurogenic and highly vascularized hippocampus and to enrich for specific vessel types. In the past, approaches to isolate endothelial cells were dependent on transgenic mice and often resulted in insufficiently pure cell populations and poor yield. This protocol describes a technique that allows single-cell isolation of highly pure brain endothelial cell populations using fluorescence activated cell sorting (FACS). Briefly, after perfusion and careful removal of the meninges, and dissection of the cortex/hippocampus, the brain tissue is mechanically homogenized and enzymatically digested resulting in a single cell suspension. Cells are stained with fluorochrome-conjugated antibodies identifying CD31+ brain endothelial cells, as well as CD45+CD11b+ myeloid cells for exclusion. Using flow cytometry, cell populations are separated and CD31+BECs are sorted in bulk into RNA later or as single cells directly into either RNA lysis buffer for single or bulk RNA-Seq analyses. The protocol does not require the expression of a transgene to label brain endothelial cells and thus, may be applied to any mouse model. In our hands, the protocol has been highly reproducible with an average yield of 1 x 105 cells isolated from an adult mouse cortex/hippocampus.
Keywords: Brain endothelial cellsBackground
The Blood-brain barrier (BBB) is a complex vascular structure that separates and protects the brain from systemic circulating factors and immune cells while allowing selective transport of critical nutrients. This highly specialized vasculature regulates selective transport of metabolites across the barrier. BECs also form a central component of the neurovascular unit (NVU), with close association and crosstalk with various cell types in the brain including neural precursor cells, microglia and the brain-resident immune cells. The BBB not only limits the passage of ions and other molecules such as glucose but also prevents the uncontrolled exchange of toxins, bacteria, viruses and cells between the blood and the brain parenchyma (Abbott et al., 2010). Nutrient-rich, oxygenated blood is pumped into the brain through cerebral arterial BECs (arteries and arterioles), which are protected and supported by smooth muscle cells (SMCs) that cover the endothelium and form a basement membrane layered by astrocytic end-feet of the brain parenchyma (Figure 1). The blood is transferred to highly specialized capillaries, which are comprised of BECs that form unique tight junctions and are wrapped by pericytes (Peric.) within the endothelial basement membrane, which is then covered by astrocytic end-feet. BBB capillaries are the site of controlled transport of fluids and solutes into the CNS. Immuno-surveillance and occasional extravasation of leukocytes (Leuk.) into the CNS parenchyma occurs at the level of postcapillary venous cells (venules and veins) the vascular segments into which blood flows after passing through the capillaries. Postcapillary Venules contain enlarged perivascular space between the endothelial and astrocytic basement membranes where occasional immune cells can reside (Banks, 2016; Engelhardt et al., 2017).
The outlined fluorescent activated cell sorting (FACS) single cell isolation method is based on previously described procedure for BEC isolation (Tam et al., 2012), that have been further developed and modified in context of a study aiming to determine molecular changes occurring in brain endothelial cells during aging and exposure to aged blood (Yousef et al., 2018). A schematic of the FACS procedure for BEC isolation and an example of cell gating are shown in Figure 2. Aging results in the upregulation of chronic inflammatory processes involving endothelial cell activation that contributes to reduced neural precursor cell activity and increased neuroinflammation in the aging brain (Yousef et al., 2018). The technique was successfully applied in a collaborative effort to investigating the effects of an aged systemic milieu on hippocampal neurogenesis and microglia activation (Yousef et al., 2018). The study highlights the role of the vascular cell adhesion molecule 1 (VCAM1) as a negative regulator of adult neurogenesis and inducer of microglial activity. VCAM1 is only expressed in a very small subpopulation of cells under baseline conditions and thus, was not sufficiently immunolabeled using conventional two-step antibody staining to perform FACS analysis. To that end, mice were stimulated systemically with Lipopolysaccharide (LPS) from Salmonella typhimurium to increase VCAM1 expression. The mice were then retro-orbitally injected with fluorescently labeled anti-mouse VCAM1, which resulted in a reliable detection of the VCAM1 positive brain endothelial cell subpopulation. The CD31+VCAM1+ BECs in LPS-stimulated mice could be used to set positive and negative gates in the flow sorter to quantitatively assess CD31+VCAM1+ expression in normal young and aged mice, or young mice treated with young or aged plasma and to isolate this rare subpopulation (Yousef et al., 2018; Figure 3). Additionally, VCAM1 is enriched in arterial and venous BECs and can be used to enrich and study these two vessel segmental populations (Vanlandewijck et al., 2018; Yousef et al., 2018).
In the last decade, several protocols for brain endothelial cell isolation have been employed, which tend to use transgenic endothelial cells labeled with fluorescent proteins such as GFP (Daneman et al., 2010; Vanlandewijck et al., 2018). Because these techniques are dependent on transgenic endothelial cell markers, they may not easily be applied to diseased or normal aged mouse models. This protocol allows the isolation of highly pure brain endothelial cell population from any mouse strain or murine animal model using papain-based enzymatic digestion using the commercial neural dissociation kit (MACS Miltenyi Biotec, Miltenyi). 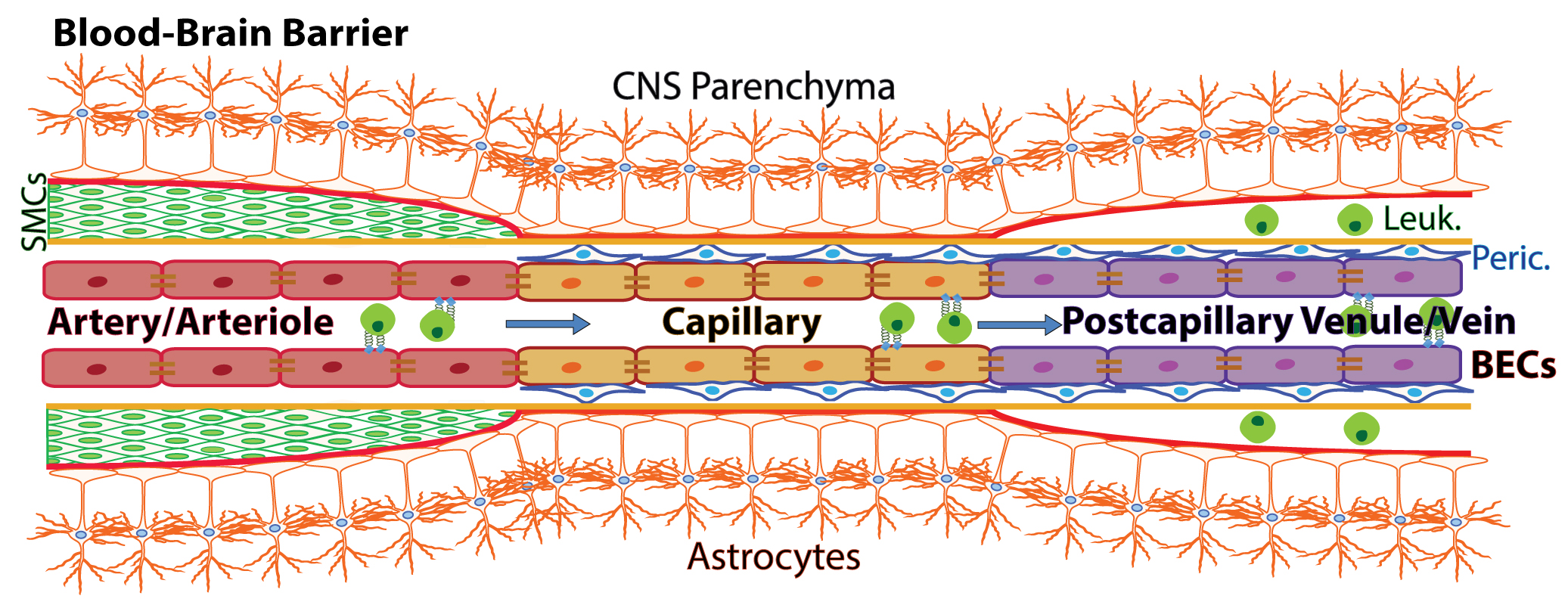
Figure 1. Schematic of the blood-brain barrier. Nutrient-rich, oxygenated blood is pumped into the brain through cerebral arterial BECs (arteries and arterioles), which are protected and supported by smooth muscle cells (SMCs) that cover the endothelium and form a basement membrane layered by astrocytic end-feet of the brain parenchyma. The blood is transferred to highly specialized capillaries, which are comprised of BECs that form unique tight junctions and are wrapped by pericytes (Peric.) within the endothelial basement membrane, which is then covered by astrocytic end-feet. BBB capillaries are the site of controlled transport of fluids and solutes into the CNS. Immuno-surveillance and occasional extravasation of leukocytes (Leuk.) into the CNS parenchyma occurs at the level of postcapillary venous cells (venules and veins) the vascular segments into which blood flows after passing through the capillaries. Postcapillary Venules contain enlarged perivascular space between the endothelial and astrocytic basement membranes where occasional immune cells can reside (Banks, 2016; Engelhardt et al., 2017). (Adopt from Yousef et al. [2018] Figure 2a)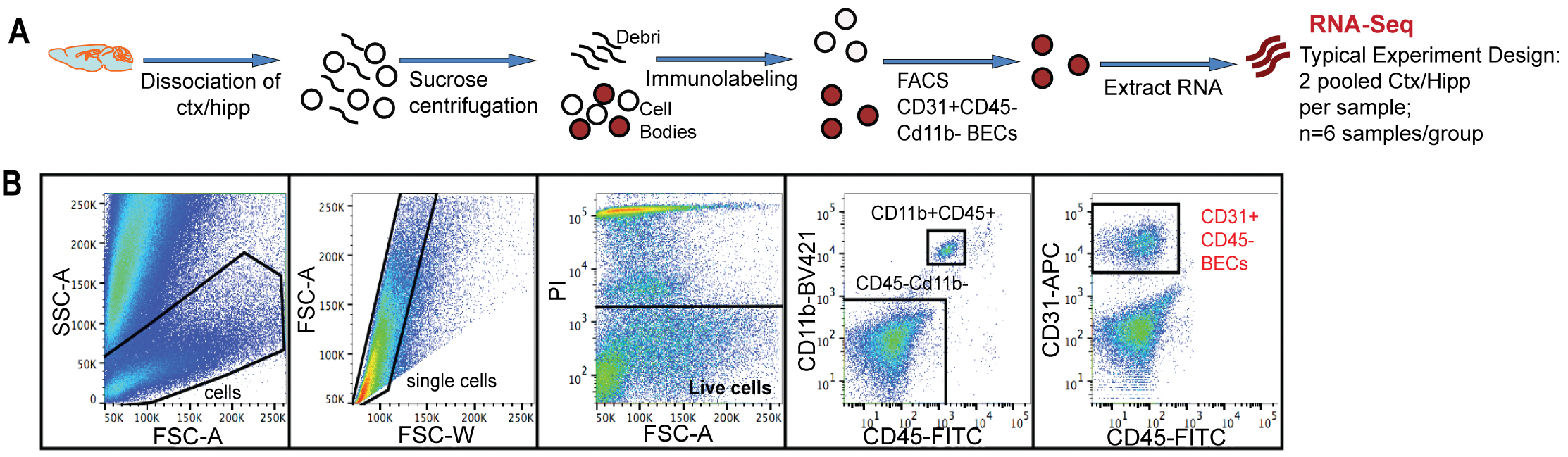
Figure 2. Mouse Brain endothelial cell isolation through flow cytometry. A. Schematic of flow sorting of CD31+CD45- BECs from mouse cortex and hippocampi. Each isolated RNA sample is a pool of BECs from 2 mouse brains. B. FACS gating strategy to isolate single BECs. PI+ dead cells were excluded. CD11b+ and CD45+ cells were gated to exclude monocytes/macrophages and microglia. CD31+Cd11b-CD45- cells were defined as the BEC population. (Adopt from Yousef et al. [2018] Supplemental Figure 1a-b)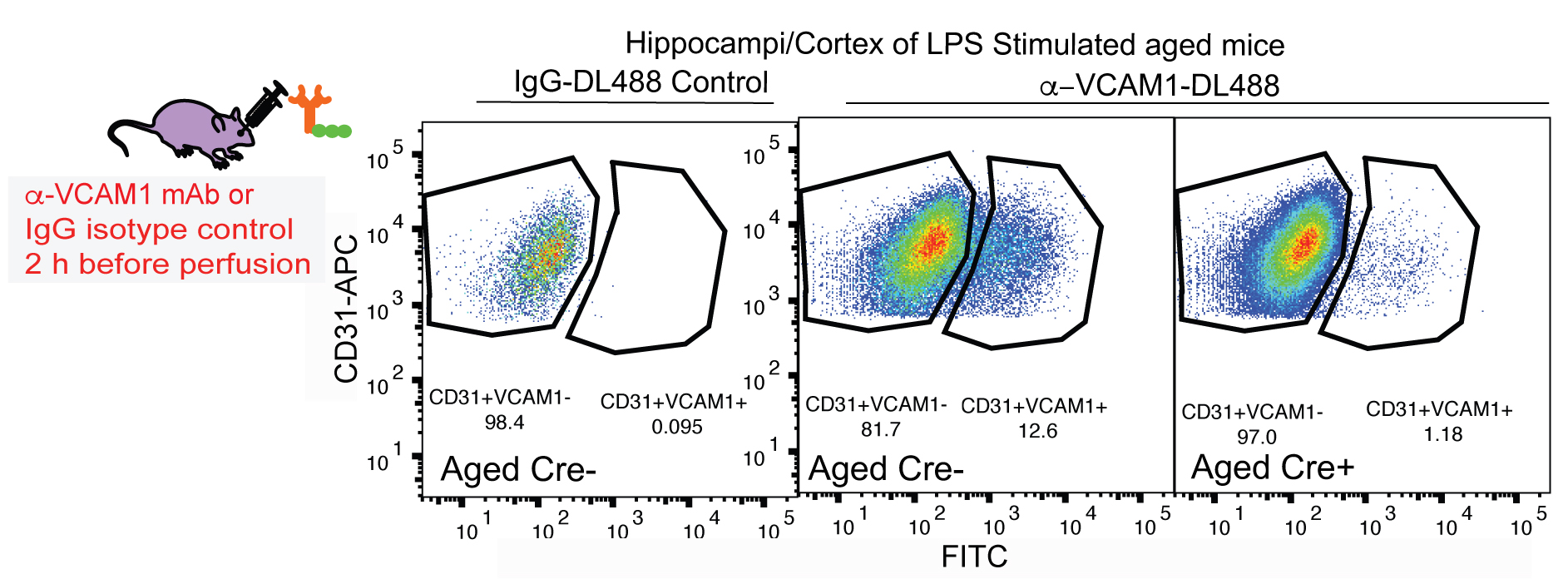
Figure 3. Identification of VCAM1+CD31+ BECs through flow cytometry. Vcam1fl/fl Slco1c1-CreERT2+/- mice (Cre+) received tamoxifen (100 mg/kg; i.p.) once daily for 5 days. After a 3-day resting period, mice were treated with LPS for 16 h and 2 h before perfusion (1 mg/kg, i.p.) and fluorescently labeled anti-VCAM1 mAb (100 µg, r.o.) for 2 h prior to cell isolation and flow cytometry analysis. Gating plots of CD31+VCAM1+ cells isolated from Gating plots are of tamoxifen-treated, LPS stimulated aged (19-month-old) Slco1c1-CreERT2+/--Vcam1fl/fl (Cre+) and littermate control mice lacking a copy of the Cre gene (Cre-), injected with fluorescently-tagged DL488 anti-VCAM1 mAb or IgG-DL488 isotype control (r.o.) 2 h before sacrifice.
Materials and Reagents
- Pipette tips must be sterile and low retention
- Whatman paper: GE Healthcare Whatman Quantitative Filter Paper Grade 40 (Fisher Scientific, GE Healthcare, catalog number: 09-927-541)
- Thermo Scientific Nalgene Rapid-Flow Sterile Disposable Filter Units with PES Membrane (Thermo Fisher, catalog number: 569-0020)
- 15 ml and 50 ml tubes (Corning, catalog numbers: 430790 and 430828)
- 1.5 ml and 2.0 ml Eppendorf Tubes Protein Low-Binding (Eppendorf, catalog numbers: 022431081 and 022431102)
- FACS Tubes: 5 ml Polystyrene Round-bottom tube with 40 μm Cell Strainer tops (Corning, Falcon®, catalog number: 352235)
- 70 μm cell strainer (Fisher Scientific, catalog number: 22-363-548)
- 40 μm cell strainer caps
- Razor blades
- Plate
- Dry ice
- 2-Mercaptoethanol (BME) (Sigma-Aldrich, catalog number: M6250-100)
- RNasin Ribonuclease Inhibitors (Promega, catalog number: N2115)
- RNA Later (Life Technologies, catalog number: AM7020)
- Miltenyi Neural Dissociation Kit (NDK) (Papain) (MACS Miltenyi Biotec, Miltenyi, catalog number: 130-092-628)
Note: Store at 2-8 °C, Shelf life: 24 months from the date of manufacture. - Fc Block: Purified Rat Anti-Mouse CD16/CD32 (Fisher Scientific, BD Pharmingen, catalog number: BDB553142) (Store at 4 °C)
- CD31-APC: APC Rat Anti-Mouse CD31 (BD Biosciences, catalog number: 551262) (Store at 2-8 °C)
- CD45-APC/Cy7: APC/Cy7 Rat Anti-Mouse CD45 (BioLegend, catalog number: 103115) (Store at 2-8 °C)
- Or CD45-FITC: FITC Rat Anti-Mouse CD45 (BD Pharmingen, catalog number: 553080, Clone 30-F11)
- CD11b-BV421: Brilliant Violet 421 Rat Anti-Mouse CD11b (BioLegend, catalog number: 101235) (Store at 2-8 °C)
- Propidium Iodide (PI): Propidium Iodide Solution (Sigma-Aldrich, catalog number: P4864) (Store at 2-8 °C)
- RNeasy Plus Micro kit (QIAGEN, catalog number: 74034)
- Optional: Rat monoclonal anti-VCAM1 (Clone M/K-2.7, Bioxell, catalog number: BE0027)
- Optional: Rat IgG1 Isotype antibody (Clone HRPN, Bioxell, catalog number: BE0088)
- Optional: DyLightTM Antibody Labeling Kit (DyLightTM 488, Thermo Scientific, catalog number: 53025)
- Modified DPBS (mDPBS) (see Recipes)
Note: Sterile filter solution, Store at 2-8 °C, Shelf life: 36 months from the date of manufacture.- Dulbecco’s Phosphate-buffered Saline (DPBS) with Calcium and Magnesium (Thermo Fisher, GibcoTM, catalog number: 14040-133)
Note: Store at 2-8 °C, Shelf life: 36 months from the date of manufacture. - Glucose (1,000 mg/L) (Fisher Scientific, ACROS Organics, catalog number: 41095-5000)
- Sodium Pyruvate (30 mg/L) (Thermo Fisher, GibcoTM, catalog number: 11360-070)
- Dulbecco’s Phosphate-buffered Saline (DPBS) with Calcium and Magnesium (Thermo Fisher, GibcoTM, catalog number: 14040-133)
- FACS buffer (see Recipes)
Note: Sterile Filter solution, Store at 2-8 °C, Shelf life: 3 months from the date of manufacture.- Modified DPBS
- 0.5% BSA (Fisher Scientific, catalog number: BP1600-100)
- 2 mM EDTA (Thermo Fisher, InvitrogenTM, catalog number: AM9912)
- 0.9 M Sucrose (see Recipes)
Sucrose in DPBS (Fisher Scientific, catalog number: BP220-10)
Note: Sterile Filter solution, Store at 2-8 °C, Shelf life: 3 months from the date of manufacture.
Equipment
- Graduated cylinder
- -80 °C freezer
- Shaker
- Centrifuge (Eppendorf, model: 5810R, catalog number: 022625004)
- Warm Water Bath (preheated at 37 °C)
- Multi-Purpose Rotator (Thermo Fisher, Lab Rotator, catalog number: 2309-1CEQ)
- BD FACSAriaTM II or III cell sorter (BD Biosciences)
- Microscissors, fine forceps
- Agilent 2100 Bioanalyzer (Agilent Technologies)
Software
- BD FACSDIVATM SOFTWARE (BD Biosciences, version: V8.0.1)
- FlowJoTM (© FlowJo, LLC, version: 9.9.4 or higher)
Procedure
Notes:
- Perform all centrifugation and staining steps at room temperature unless otherwise stated. Flow cytometry sorter should be set to 100 μm nozzle.
- Prepare all reagents ahead of time and warm to room temperature if necessary.
- Optional: To label adhesion molecules, such as VCAM1 which is uniquely enriched in arterial and venous BECs (Vanlandewijck et al., 2018; Yousef et al., 2018), on BECs for vessel segmental enrichment. If VCAM1 (or another adhesion molecule) labeling is desired, conjugate anti-VCAM1 mAb and rat IgG1 isotype control antibody to DyLightTM-488 according to manufacturer’s instructions.
- Mice: Mice were anesthetized with avertin and perfused with 20 ml cold PBS following blood collection.
- Optional: Set controls using LPS-stimulated mice: Mice were treated with LPS for 16 h and 2 h before perfusion (1 mg/kg, i.p.). Mice were injected with fluorescently labeled anti-VCAM1 mAb (100 µg, r.o.) or IgG1 isotype control 2 h prior to cell isolation and flow cytometry analysis.
- All experimental (healthy) mice are also injected with anti-VCAM1 mAb (100 µg, r.o.) 2 h prior to cell isolation if vessel segmental enrichment is desired.
- Tissue Dissociation
- For each sample, usually comprising of dissected cortex/hippocampus from 1-2 brains for applications such as bulk RNA-Seq, or pooled hippocampi (from 4 mice) for applications such as single cell RNA-Seq (Yousef et al., 2018), preheat 1.5 ml of Buffer X from Neural Dissociation Kit (NDK) + 9 μl of 2-Mercaptoethanol (BME) + 50 μl Enzyme P from NDK in 15 ml Falcon tubes in a 37 °C water bath.
- Remove meninges by rolling whole brain on Whatman paper.
- Dissect hippocampus, cortex, and remainder of the brain using microscissors and forceps in a sterile plate. Separate individual tissues and roughly chop into fine bits using a razor blade. Chop the brain segments to such a degree that you can pass the minced tissue through a trimmed, 1 ml pipettor (trimmed meaning the very tip is cut off to increase the diameter of the hole so that minced tissue can get through), but not so chopped that it becomes very mushy and could go through an untrimmed 1 ml pipettor. If the brain is too minced, it might be overly digested in the enzyme mixture in subsequent steps leading to cell loss.
- Alternatively, if isolating the hippocampus or other very small brain regions separately, it is not necessary to mince in a sterile plate. Rather, transfer each dissected small tissue using sterile forceps into 1.5 ml Eppendorf tubes with 0.5 ml of Buffer X from NDK and chop finely with microscissors.
- Using a trimmed 1 ml tip (trimmed meaning the very tip is cut off to slightly increase the diameter of the hole), transfer finely chopped samples to the preheated Buffer X solution prepared in Step A1. Triturate 10 times with the same pipette tip. Incubate in a 37 °C water bath for 15 min. Flick tubes every 5 min of incubation. Prepare Enzyme 2 Mix from NDK for each sample: 10 μl of Enzyme A + 20 μl Buffer Y.
- Add 30 μl of Enzyme 2 Mix into each sample and triturate 10 times with a 1 ml pipette tip, start 10 min timer from when the first sample receives the Enzyme 2 Mix. Incubate samples in a 37 °C water bath for the remainder of the 10 min timer, flick samples every 5 min.
Note: Be quick! Do not over incubate, over digestion will result in poor yields. - Prepare 50 ml tubes with 70 μm cell strainers, wash strainers with 1 ml of mDPBS. Collect the flowthrough in the 50 ml tube.
- Triturate samples 10 times with a 1 ml pipette tip and immediately add 10 ml mDPBS.
- Pass all of the supernatant through a 70 μm cell strainer prepared in Step A7. Wash through another 5 ml mDPBS.
- Centrifuge at 300 x g for 10 min. Discard the supernatant and collect the pellet. Carefully pipette out supernatant without disturbing the pellet.
- Myelin Removal and Staining
- Resuspend pellets in 5 ml (2.5 ml for Hippocampus samples) of 0.9 M Sucrose and transfer each pellet to a 15 ml tube. Centrifuge at 850 x g for 15 min. Discard the supernatant, and collect the pellet. Carefully pipette out and discard the supernatant without disturbing the pellet.
- Sample resuspension
- For Hippocampus samples:
Resuspend pellet in 2 ml FACS buffer, transfer to a 2 ml Eppendorf tube and wait for other tissue type samples before continuing to Step B3. - For Cortex and Whole Brain samples:
Resuspend pellet in 1 ml 0.9 M Sucrose before further adding another 2 ml of Sucrose. Centrifuge at 850 x g for 15 min. Discard supernatant without disturbing the pellet. Resuspend the pellet in 2 ml FACS buffer, transfer to a 2 ml Eppendorf tube.
- For Hippocampus samples:
- Centrifuge all samples at 300 x g for 5 min. Prepare Fc Blocking solution (100 µl per sample):
For 10 samples, dilute 10 μl Fc Block in 1 ml FACS buffer. Plan which Compensation standards will be needed (Single Channel comps for each marker, and 2 unstained controls, with and without propidium iodide (PI) which is added in Step C3 below). Cell pellets are visible for cortex pooled samples, but likely not visible for hippocampal samples. Note the estimated cell numbers in Step C6 below. - Resuspend each sample in 100 μl of Fc Blocking solution prepared in Step B3 above (For the sample to be used in Compensation standards, add an extra 25 μl of Fc Blocking solution to the sample for each planned Compensation standard). Incubate on a shaker for 5-10 min at room temperature.
- Add the following antibodies (1:100) and incubate on a shaker for 30 min at RT:
Anti-CD31-APC
Anti-CD11b-BV421
Anti-CD45-APC/Cy7 or anti-CD45-FITC
Note: If mice were injected with fluorescently labeled anti-mouse VCAM1-DyLightTM488 as described above, stain CD45 in the APC/Cy7 channel, and also gate CD31+VCAM1+ cells in the APC and FITC channels. - Add 1.8 ml of FACS buffer to dilute the samples, then centrifuge at 300 x g for 5 min.
- Prepare Sorting Mix: FACS buffer + PI (1:5,000 of a 1 mg/ml stock) + RNase Inhibitor (1:500) (For 5 ml: Add 1 μl PI and 10 μl RNase Inhibitor).
- Carefully pipette out supernatant and resuspend each sample (except Compensation standards, resuspend comps in 0.5 ml FACS buffer) in 0.5 ml of Sorting Mix.
- Transfer all samples through cell strainer capped FACS tubes (Blue 40 μm cell strainer caps). Keep on ice and covered from light until sorted by flow cytometry.
- If collecting whole population cells: Prepare 1.5 ml RNase-free collection tubes with 0.5 ml of RNA later. Be prepared to dilute samples to a total volume of 1.3 ml with RNA later after collecting cells and snap freeze samples with dry ice.
- If collecting single cells: have plates prepared with RNA lysis buffer as instructed by manufacturer for the technique chosen, and kept frozen and stored at -80 °C until used for cell sorting (aka, Smart-seq-2 protocol as described previously [Picelli et al., 2014; Darmanis et al., 2015], was used in Yousef et al. [2018] study).
- This protocol does not cover detailed instructions for FACS parameter setup. For assistance, please contact your FACS core facility manager or FACS machine vendor.
- Use a 100 µm nozzle.
- While sorting, keep the threshold rate around or below 100x the value of the set frequency (typically fewer than 2,800 events per second).
- Estimated sort time is 15-20 min per stained sample.
- FACS and sample collection
- Run unstained cells to set up forward (FSC-A) and side scatter (SSC-A) (Figure 2B).
- Run single-color controls to set FACS parameters and compensate for channel spillover.
- Add 1:5,000 PI from a stock of 1 mg/ml (0.2 µg/ml final concentration) (or 1:500 from 1:10 pre-dilution) PI for live/dead staining right before running the individual sample, if it was not already added at Step B7 of Myelin Removal and Staining section above.
- For your first run and whenever obtaining a new antibody: Run controls in which cells are stained with all antibodies except for one, to determine positive cell populations and set up gates accordingly.
- Run stained FACS sample:
- Gate cells on forward (FSC-A = size) and sideward scatter (SSC-A = internal structure) to exclude cell debris and residual myelin
- Plot FSC-A against FSC-W to discriminate single cells from cell doublets/aggregates.
- Exclude PI positive (dead) cells.
- Exclude CD11b+CD45+ monocytes/macrophages and microglial cells by gating on CD45 and CD11b negative cells.
- Gate on CD31+ BECs and collect cells from this gate to obtain a pure brain endothelial cell population. Record at least 1 x 106 events.
- Collect cells in either:
- 1.5 ml RNase-free tubes containing FACS buffer (if RNA integrity is less of a concern) or 0.5 ml RNA later (to preserve for later RNA isolation for bulk RNA-Seq). If containing 0.5 ml of RNA later:
- Ideally, 100,000 cells or more are needed for optimal RNA extraction. Top off the RNA later up to 1.3 ml before mixing well and snap freezing the samples with dry ice. Store at -80 °C until RNA isolation.
- When spinning down the cells to extract RNA, thaw cells and warm to room temperature before 10 min centrifugation at 1,000 x g. Isolate total RNA from the cell pellets using the RNeasy Plus Micro kit (QIAGEN). Assess RNA quantities and RNA quality using the Agilent 2100 Bioanalyzer (Agilent Technologies). All samples passed a quality control threshold (RIN ≥ 8.5) to proceed to library preparations and RNA-Seq.
- Single cells in a 96-well plate with RNA lysis buffer for single cell RNA-Seq (according to Smart-Seq-2 [Picelli et al., 2014; Darmanis et al., 2015] or other preferred protocol).
Notes:- Keep plates frozen on dry ice until just before they are used to sort cells. Immediately after cell sorting, cover each plate with an aluminum freezing compatible lid, vortex the plate to mix cells with RNA lysis buffer, and freeze plate on dry ice until it can be transferred and stored at -80 °C until RNA isolation.
- For a typical sample of 1-2 cortex/hippocampi to isolate cells in bulk for RNA extraction (aka for bulk RNA-Seq), sorting by flow cytometry will result in around 100,000 isolated CD31+BECs per sample. For a typical sample of 4 pooled hippocampi, sorting by flow cytometry results in collection of a few thousand to tens of thousands of cells which varies by prep, cell collection method (bulk or single cell) and if VCAM1 is enriched.
- 1.5 ml RNase-free tubes containing FACS buffer (if RNA integrity is less of a concern) or 0.5 ml RNA later (to preserve for later RNA isolation for bulk RNA-Seq). If containing 0.5 ml of RNA later:
Data analysis
- Please refer to Yousef et al. (2018) published in BioRxiv for the data analysis
This BEC isolation protocol was used in the following figures in the Yousef et al. (2018) paper:
Figure 1a-c: Bulk RNA-Seq of young and aged cortex/hippocampal BECs.
Supplemental Figure 1a-e: Bulk RNA-Seq of young and aged cortex/hippocampal BECs.
Supplemental Figure 1f-k: C57BL6 mice were injected with anti-VCAM1-DL488 or IgG-DL488 isotype control (r.o.) 2 h before perfusion to label BECs in vivo prior to brain dissociation, staining, and FACS. The hippocampal cells were then used for single cell RNA-Seq (Figure 2 and Supplemental Figure 2).
Figure 2 and Supplemental Figure 2: Single cell RNA-Seq of BECs isolated from the hippocampus.
Figure 3c-d: BECs isolated from young mice treated with young or aged plasma and injected with anti-VCAM1-DL488 or IgG-DL488 isotype control (r.o.) 2 h before perfusion to label BECs in vivo prior to brain dissociation, staining, and FACS. Young mice stimulated with LPS were used to set the gates. - Please refer to “Collagenase-based Single Cell Isolation of Primary Murine Brain Endothelial Cells Using Flow Cytometry” by Czupalla et al. (2018) Bio-Protocol, for an alternative protocol for BEC cell isolation by flow cytometry which uses a more gentle collagenase-based method and several markers to stain additional cell types. This protocol was used for Supplemental Figure 7a-d in the Yousef et al. (2018) paper published in BioRxiv to show genetic recombination and deletion of Vcam1 in tamoxifen-treated Vcam1fl/flSlco1c1-CreERT2+/- mice.
Recipes
- Modified DPBS (mDPBS) (500 ml)
- Measure 500 mg of glucose in a graduated cylinder
- Add 1.36 ml of 100 mM sodium pyruvate to the graduated cylinder, and then add DPBS up to the 500 ml notch
- Seal the graduated cylinder using parafilm and mix thoroughly by inverting several times
- Once mixed, sterile filter using a Thermo Scientific Nalgene Rapid-Flow Sterile filter unit with 0.2 μm pore
- FACS buffer (500 ml)
- Add 2.5 g of BSA and 2 ml of 0.5 M EDTA to 500 ml of mDPBS
- Mix thoroughly and sterile filter using a Thermo Scientific Nalgene Rapid-Flow Sterile filter unit with 0.2 μm pore
- 0.9 M Sucrose (500 ml)
- Measure 154.04 g of sucrose in a graduated cylinder
- Add DPBS up to the 500 ml notch
- Seal the graduated cylinder using parafilm and mix thoroughly by inverting several times
Acknowledgments
We thank Lusijah Sutherland PhD, and Corey Cain PhD, for managing the core flow cytometry facility at the VA in Palo Alto and providing H.Y. and C.J.C. training on the instruments; Corey Cain PhD as well for his experimental advice, assistance with flow cytometry and analysis of PBMCs and thoughtful discussion. We would also like to thank Ryan Watts PhD and Nga Bien-Ly PhD for sharing the original BEC isolation protocol used for RNA-Seq. This work was funded by the Department of Veterans Affairs (T.W.-C.), the National Institute on Aging (SPO: 116650; 1F32AG051330-01A1 to H.Y., R01-AG045034 and DP1-AG053015 to T.W.-C.), the NOMIS Foundation (T.W.-C.), The Glenn Foundation for Aging Research (T.W.-C), a SPARK grant to H.Y. through the Stanford Clinical and Translational Science Award (CTSA) to Spectrum (UL1 TR001085), the National Institutes of Health (R01-GM37734 and R37-AI047822 to E.C.B, RO1 AI109452 to HH), the Stanford Institute for Immunity, Transplantation and Infection (C.J.C.), and the Edinger Institute (C.J.C.). The CTSA program is led by the National Center for Advancing Translational Sciences (NCATS) at the National Institutes of Health (NIH).
Competing interests
There are no competing conflicts of interest.
Ethics
The mice used in this protocol were bred and aged in-house and lived under a 12 h light-dark cycle in pathogenic-free conditions, in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health.
References
- Abbott, N. J., Patabendige, A. A., Dolman, D. E., Yusof, S. R. and Begley, D. J. (2010). Structure and function of the blood-brain barrier. Neurobiol Dis 37(1): 13-25.
- Banks, W. A. (2016). From blood-brain barrier to blood-brain interface: new opportunities for CNS drug delivery. Nat Rev Drug Discov 15(4): 275-292.
- Czupalla, C. J., Yousef, H., Wyss-Coray, T. and Butcher, E. C. (2018). Collagenase-based single cell isolation of primary murine brain endothelial cells using flow cytometry. Bio-protocol 8(22): e3092.
- Daneman, R., Zhou, L., Agalliu, D., Cahoy, J. D., Kaushal, A. and Barres, B. A. (2010). The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One 5(10): e13741.
- Darmanis, S., Sloan, S. A., Zhang, Y., Enge, M., Caneda, C., Shuer, L. M., Hayden Gephart, M. G., Barres, B. A. and Quake, S. R. (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112(23): 7285-7290.
- Engelhardt, B., Vajkoczy, P. and Weller, R. O. (2017). The movers and shapers in immune privilege of the CNS. Nat Immunol 18(2): 123-131.
- Picelli, S., Faridani, O. R., Bjorklund, A. K., Winberg, G., Sagasser, S. and Sandberg, R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9(1): 171-181.
- Tam, S. J., Richmond, D. L., Kaminker, J. S., Modrusan, Z., Martin-McNulty, B., Cao, T. C., Weimer, R. M., Carano, R. A., van Bruggen, N. and Watts, R. J. (2012). Death receptors DR6 and TROY regulate brain vascular development. Dev Cell 22(2): 403-417.
- Vanlandewijck, M., He, L., Mae, M. A., Andrae, J., Ando, K., Del Gaudio, F., Nahar, K., Lebouvier, T., Lavina, B., Gouveia, L., Sun, Y., Raschperger, E., Rasanen, M., Zarb, Y., Mochizuki, N., Keller, A., Lendahl, U. and Betsholtz, C. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554(7693): 475-480.
- Yousef, H., Czupalla, C. J., Lee, D., Burke, A., Chen, M., Zandstra, J., Berber, E., Lehallier, B., Mathur, V., Nair, R. V., Bonanno, L., Merkel, T., Schwaninger, M., Quake, S., Butcher, E. C. and Wyss-Coray, T. (2018). Aged blood inhibits hippocampal neurogenesis and activates microglia through VCAM1 at the blood-brain barrier. bioRxiv. doi:10.1101/242198.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yousef, H., Czupalla, C. J., Lee, D., Butcher, E. C. and Wyss-Coray, T. (2018). Papain-based Single Cell Isolation of Primary Murine Brain Endothelial Cells Using Flow Cytometry. Bio-protocol 8(22): e3091. DOI: 10.21769/BioProtoc.3091.
Category
Neuroscience > Cellular mechanisms > Cell isolation and culture
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









