- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ex vivo Culture and Lentiviral Transduction of Benign Prostatic Hyperplasia (BPH) Samples
Published: Vol 8, Iss 21, Nov 5, 2018 DOI: 10.21769/BioProtoc.3075 Views: 6160
Reviewed by: Tomas AparicioAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Three-dimensional Models of the Nasopharynx for the Study of Epstein-Barr Virus Infection
Phillip Ziegler [...] Kathy Ho Yen Shair
Mar 20, 2022 4035 Views

High-Efficiency Retroviral Transduction for Type 1 Regulatory T Cell Differentiation
Michael C. McGee [...] Weishan Huang
Sep 5, 2022 2697 Views

Human Skin Explant Preparation and Culture
Jessica L. Shannon [...] Jennifer Y. Zhang
Sep 20, 2022 3956 Views
Abstract
To assess oncogenic potential, classical transformation assays are based on cell line models. However, cell line based models do not reflect the complexity of human tissues. We thus developed an inducible expression system for gene expression in ex vivo human tissues, which maintain native tissue architecture, such as epithelia and stroma. To validate the system, we transduced and expressed known tumor suppressors (p53, p33ING1b), oncoproteins (RasV12, p47ING3), or controls (empty vector, YFP) in ex vivo prostate tissues, then assessed proliferation by immunohistochemistry of markers (H3S10phos). Herein, we describe how to generate lentiviral vectors and particules, successfully transduce human prostate tissues, induce exogenous gene expression, and assess cellular proliferation.
Keywords: Ex vivo tissue culture modelBackground
Cancer afflicts all classes of society, but incidence rates increase with age. Thus, with increasing lifespan in developed countries, cancer poses an increasing burden on healthcare systems. Most investigations rely on cancer cell lines, which have defined phenotypes and genotypes, are simple to maintain in the laboratory, and usually straightforward methods are established to manipulate them (e.g., introduce genes, silence genes…). However, cells do not exist as individuals in the body, they are an integral part of tissues, which are vascularized and composed of various cell types that make cell-cell contacts and exchange signaling molecules. Thus, tissues are evidently more representative and relevant models than individual cells to study cell biology and pathologies, such as cancer.
Although several factors are involved in cancer development, such as unrepaired DNA damages, reactive oxygen species, metabolism, genomic instability, the principal model remains intact. Specifically, tumor suppressors are either silenced or mutated while oncoproteins get activated, leading to uncontrolled proliferation and cancer development.
While various cellular proliferation assays (cell counting, transformation, clonogenic, live cell imaging…) and gene expression manipulation methods are well established for cell lines, comparably reliable methods are lacking in the nascent field of ex vivo tissue models. We thus established a robust method for lentiviral transduction and inducible gene expression in ex vivo prostate tissues.
Prostate cancer (PC) is the most common UK male cancer. Most patients respond to androgen ablation therapy that targets the androgen receptor (AR), but patients invariably progress to castrate resistant PC (CRPC) for which there is no curative treatment available and where the prognosis is extremely poor (Feldman et al., 2001). Research investigating novel therapeutics and drug efficacy in PC is hampered by the lack of reliable models that reproduce the patients’ disease. Few PC cell lines are available and they only represent very late disease. Established, well-characterized cell lines (LNCaP, VCaP, DUCaP, CWR22R, PC3 and DU145) were developed from metastasis with AR mutation, amplification and AR-variant expression or even AR loss, but are not representative of early disease presentation for the majority of men where AR gene is generally unaltered (Reference 1). Additionally, cells are cultured on plastic as 2D monolayers of a homogeneous cell population and therefore lack tissue architecture and interactions of distinct cell types. The importance of the tumor microenvironment is also increasingly acknowledged with not only epithelium, but also stromal components (not represented in cell lines), which contribute to PC and CRPC development. Clinical value of using genetically engineered mouse models has the caveat that mice fail to develop PC and their prostate anatomy is dramatically different from the human prostate. Moreover, animal xenograft experiments suffer similar limitations as cell line-based investigations and are further limited by most xenografts being subcutaneously implanted and do not accurately represent complex, heterogeneous tumors. Not surprisingly therefore, there is high variability in the correlation between xenograft studies and clinical trials reported in PC (Centenera et al., 2012 and 2013). These factors substantially contribute towards the very low success rate observed in phase III clinical trials of novel PC therapeutics (significantly less than the general cancer average of 5%). Identification of new, more clinically relevant models of PC that are specific to individual PC patients would allow to improve this statistic and to address an unmet need. Thus, we have developed an ex vivo prostate tissue model to assess proliferation, but also oncogenic and tumor suppressive potential of exogenously expressed genes. These cultures can be generated from patients at all stages of disease and maintain tissue architecture, comprising both stroma and epithelial compartments, thereby retaining crucial contributory autocrine and paracrine signaling interactions.
Materials and Reagents
- Lentiviral vector generation
- pLVX Lenti-X Tet-One inducible expression system (Clontech, catalog number: 631847)
- FWDseq: taaaccagggcgcctataaa (IDT custom primer)
- REVseq: taggcagtagctctgacggc (IDT custom primer)
- BamHI (NEB, catalog number: R3136S)
- EcoRI (NEB, catalog number: R3101S)
- Calf Intestinal Alkaline Phosphatase (CIAP; NEB, catalog number: M0290S)
- Hi-fidelity DNA polymerase for PCR amplification (e.g., Platinum PCR SuperMix, Invitrogen, catalog number: 12532-016 or PfuTurbo DNA Polymerase, Agilent Technologies, catalog number: 600252)
- In-Fusion HD enzyme (Clontech, catalog number: 638910)
- QIAquick gel extraction columns (QIAGEN, catalog number: 28704)
- Ampicillin 100 mg/ml (dissolved in ddH2O and filtered sterilized)
- Small-scale plasmid DNA purification QIAprep Spin Miniprep kit (QIAGEN, catalog number: 27104)
- Large-scale plasmid DNA purification Maxi kit (QIAGEN, catalog number: 12163)
- Yeast extract
- Tryptone
- NaCl
- 2YT media (see Recipes)
- Lentiviral particules packaging
- Syringe
- 0.45 μm filters
Caution: Make sure to use low protein-binding, such as Millex-HV Filter PVDF (Millipore, catalog number: SLHVM33RS) to prevent binding of viral particules to the membrane and loss. - 100 mm culture dishes
- Amicon Ultra-15 (Millipore, catalog number: UFC903024)
- HEK293T cell line (ATCC, catalog number: CRL-3216)
- Packaging plasmids (pMD2.G and psPAX2 from Addgene, catalog numbers: 12260 and 12259, respectively) to be transformed and purified (e.g., Maxi kit, QIAGEN, catalog number: 12163)
- Transfection reagent TransIT-LT1 (Mirus MIR, catalog number: 2305)
- Dulbecco's Modified Eagle's Medium (DMEM, Sigma-Aldrich, catalog number: D6171)
- Lentiviral particules transduction and titration
- 6-well culture plates
- LNCaP cell line (ATCC, catalog number: CRL-1740)
- RPMI-1640 media (Sigma-Aldrich, catalog number: R5886)
- 200 mM L-glutamine (Invitrogen)
- Fetal calf serum (Gibco)
- Hexadimethrine bromide (Polybrene, Sigma-Aldrich, catalog number: H9268)
- Puromycin dihydrochloride (Sigma-Aldrich, catalog number: P9620)
- Crystal violet
- Induction of gene expression
- 10% ethanol
- Monoclonal ANTI-FLAG M2-Peroxidase (HRP) antibody (Sigma-Aldrich, catalog number: A8592)
- Doxycycline (Sigma-Aldrich, 10 mg/ml)
- 4% formalin
- Ex vivo tissue preparation and transduction
- Scalpels or surgical blades
- Hydrophobic pen (DAKO)
- 100% ethanol
- Media (RPMI-1640, Sigma-Aldrich, catalog number: R5886)
- Gelatin sponges (Spongostan, Johnson & Johnson, catalog number: 1364245)
- Antimycotic solution (Sigma-Aldrich, catalog number: A5955)
- 10 μg/ml hydrocortisone (Sigma-Aldrich)
- 10 μg/ml insulin solution from bovine pancreas (Sigma-Aldrich, catalog number: I0516)
- Hexadimethrine bromide (Polybrene, Sigma-Aldrich, catalog number: H9268)
- Doxycycline (10 mg/ml)
- Bovine Serum Albumin (Sigma-Aldrich)
- HRP-conjugated secondary antibodies
ImmPRESSTM HRP Anti-Rabbit IgG (Vector Laboratories, catalog number: MP-7401)
ImmPRESSTM HRP Anti-mouse IgG (Vector Laboratories, catalog number: MP-7402) - ImmPACT DAB Peroxidase (HRP) Substrate (Vector Laboratories, catalog number: SK-4105)
- Xylene
- DPX mounting media (Sigma)
- Tetracycline-free FBS (Clontech, catalog number: 631106)
- Immunohistochemistry staining for proliferation markers
- Primary antibodies for proliferation markers: H3S10phos (Millipore, catalog number: 06-570)
- Antigen Retrieval Buffer (100x Citrate Buffer pH 6.0) (e.g., Abcam, catalog number: ab93678)
- NaHCO3
- MgSO4
- Hematoxylin
- Scott's tap water substitute (see Recipes)
- Gill's Hematoxylin II solution (see Recipes)
Equipment
- Pipettes (e.g., Gilson)
- PCR machine
- CO2 incubator
- Decloaker
- Microtome to slice paraffin blocks
- Freezer
- Shaker
Software
- Aperio imaging system (https://www.leicabiosystems.com/digital-pathology/scan/)
Procedure
- Lentiviral vector generation (Timing 1-2 weeks)
- Amplify by PCR the gene(s) of interest to insert in pLVX using gene-specific primers:
- Fwd ccctcgtaaaGAATTC xxx xxx xxx xxx xxx xxx xxx
- Fwd cgactacgccGGATCC xxx xxx xxx xxx xxx xxx xxx if using pLVX-FLAG/HA (McClurg et al., 2018)
- Rev ccctcgtaaaGAATTC yyy yyy yyy yyy yyy yyy yyy
- Assemble PCR reaction:
Template DNA 0.1 μg
Platinum PCR SuperMix 45 μl
2 μl of Fwd primer 10 μM
2 μl of Rev primer 10 μM
Add ddH2O to a final volume of 50 μl - Run PCR programme:
Denature 5 min at 95 °C
25 cycles:
Denature - 30 s at 95 °C
Anneal - 30 s at 55 °C
Extend - 1 min per 1,000 bp of PCR product at 68 °C
Final extension at 68 °C
Ramp down to 4 °C and keep at 4 °C indefinitely
Pause Point: The PCR product(s) can be kept for several months at -20 °C. (see Table 1-[1]) - Linearise pLVX (or pLVX-FLAG/HA available upon request) by digesting 2-10 μg purified plasmid with BamHI and EcoRI (or only BamHI if using pLVX-FLAG/HA) for 30-60 min at 37 °C, following instructions from manufacturer (see Michael and Joseph [2012], if more background is required for this or following steps). Then, gel purify by running on 1% agarose gel, cutting the ~9,300 bp fragment, dissolve in 600 μl QIAGEN solubilization buffer, apply on column, spin for 1 min at maximum speed, remove filtrate, wash with 700 μl QIAGEN wash buffer, spin for 1 min at maximum speed, remove filtrate, spin one last time for 1 min at maximum speed, apply 50 μl 50 μM Tris-Cl pH 7.5 elution buffer, let sit for 2-5 min, spin for 1 min at maximum speed, remove column, and retain purified pLVX linearised plasmid DNA.
Pause Point: The linearised pLVX DNA can be kept for several months at -20 °C. - Insert the PCR product into the pLVX vector by recombination with In-Fusion HD enzyme (following the manufacturer's instructions).
- Transform competent DH5α bacterial cells using standard procedures.
- Purify DNA using QIAGEN QIAprep Spin Miniprep kit
Caution: Make sure constructs are verified by sequencing using gene-specific primers if necessary as well as pLVX FWDseq (taaaccagggcgcctataaa) and REVseq (taggcagtagctctgacggc) primers.
Pause Point: The purified plasmids can be stored indefinitely at -20 °C until ready. - Transform positive clones in DH5α. Pick a colony in the morning, inoculated 2 ml 2YT media (supplemented with 100 μg/ml ampicillin), incubate in a shaker (200-250 rpm) at 37 °C for about 6-8 h, then scale up to 250 ml 2YT media (supplemented with 100 μg/ml ampicillin), and return to shaker (200-250 rpm) at 37 °C overnight.
- Purify plasmid DNA using QIAGEN Maxi kit according to the manufacturer’s instructions
Pause Point: The purified plasmids can be stored indefinitely at -20 °C until ready to package lentiviral particules.
- Amplify by PCR the gene(s) of interest to insert in pLVX using gene-specific primers:
- Lentiviral particules packaging (Timing 5 days)
- The day before transfection, seed 2-3 million HEK293T cells in 100 mm dishes (1 for each pLVX construct) in 10 ml complete DMEM. Too few or too many cells will result in poor transfection and low lentiviral particle yields.
- The next day, prepare for each pLVX construct to transfect, 1 ml of serum-free media and 54 μl of LT1 reagent. Incubate the media-LT1 mixture at room temperature for 15 min. During that time, mix 9 μg pLVX with 6.75 μg psPAX2 and 2.25 μg pMD2.G. Add 1 ml of the media-LT1 mixture to the DNA and further incubate for 30 min at room temperature. Retrieve the HEK293T dishes from the incubator and add the media-LT1-DNA mixture dropwise to the cells. Return the HEK293T dishes to the incubator and leave overnight (12-18 h). The next day, remove the media (~10 ml) and replace with 5-6 ml of fresh DMEM (with supplements such as FBS, glutamine, and antibiotics).
- After 24 h and 48 h, collect the media (which contains the lentiviral particules - viral supernatant), replenish with 5-6 ml fresh DMEM and store in a refrigerator. Once the viral supernatants are collected, the viruses should be filtered (essentially to remove floating HEK293T cells) using a syringe and a 0.45 μm filter (Millex-HV Filter PVDF; Millipore).
Caution: Make sure the specified low protein-binding 0.45 μm filters are used, otherwise the viral particules may bind to the filter, reducing drastically the viral titre.
Pause Point: The viral particules can be stored in the refrigerator for several months, aliquoted and stored at -20 or -80 °C, or concentrated and stored at -80 °C.
- Lentiviral transduction and titration (Timing 5 days)
- On Day 0, seed LNCaP cells at 100,000 cells per well of a 6-well plate.
- On Day 1, add lentiviral particules to each well following a serial dilution scheme [e.g., 2,000 μl (dilution factor 1), 200 μl (10), 20 μl (1 x 102), 2 μl (1 x 103), 0.2 μl (1 x 104), and a 0 μl not transduced control]. The viral particules are left on the cells overnight (about 18 h).
- On Day 2, remove the lentivirus-containing media and replace with fresh complete RPMI-1640 media.
- On Day 3, add puromycin to each well at a 1 μg/μl final concentration.
- On Day 5, remove the media and fix and stain the cells using 1% crystal violet dissolved in 10% ethanol (enough to cover the plate). Wash the stain several times with distilled water and count colonies. The multiplicity of infection (MOI) is estimated using the following formula:
MOI = colony number X viral dilution factor (see Table 1-[2])
Caution: Try to avoid freeze-thawing the lentiviral stocks as this may reduce the MOI. Of note, we observed that the lentiviral supernatants are relatively stable for several months at 4 °C without major loss of MOI.
- Ex vivo tissue preparation and transduction (Timing 1-2 weeks)
- Benign prostatic hyperplasia (BPH) samples are obtained from cancer-free patients according to local regulations and with ethical approval from relevant entities. BPH samples are removed surgically following patient’s consent and placed in enough ice cold RPMI1640 media without supplements to cover the specimen.
- Within 24 h from the surgery, dissect tissues to 1 mm3 pieces and culture in duplicates on gelatin sponges (Spongostan, Johnson & Johnson) pre-soaked in culture media supplemented with 1x antimycotic solution (Sigma), 10 μg/ml hydrocortisone, and 10 μg/ml insulin solution from bovine pancreas (Sigma), doxycycline, and lentiviral particules.
- At the termination of the experiments (24-72 h), immediately place the samples in 4% formalin for 24 h, then process in ethanol, followed by xylene, and finally paraffin embedding.
Pause Point: Once fixed and embedded in paraffin, the ex vivo tissues can be stored at room temperature indefinitely.
Caution: It is critical that lentiviral particles containing media and doxycycline are added to the culture media at the stage of presoaking the sponges to ensure that they penetrate through the sponge and reach the explant. (see Table 1-[3])
- Immunohistochemistry staining for proliferation markers (Timing 2 days)
- Slice formalin fixed paraffin embedded tissues and then bake at 60 °C for 2 h.
- Deparaffinize the tissues by incubation in three consecutive pots of xylene for 5 min each.
- Hydrate the tissues by sequential incubation for 1 min in 100% ethanol three times followed by 95% ethanol, 70% ethanol, 50% ethanol, and finally water.
- Retrieve antigens in citrate buffer in a decloaker at 125 °C for 30 s.
- Inactivate endogenous peroxidase by incubating cooled slides in 3% H2O2 solution for 10 min, then wash slides in water and block non-specific protein binding sites by a 10 min incubation in 1% BSA in TBS. Use a hydrophobic pen (DAKO) to mark around the tissue.
- Incubate the slides in primary antibody diluted in 1% BSA 200 µl per section overnight at 4 °C in a wet chamber.
- Wash the slides twice in TBS for 5 min with gentle agitation.
- Incubate the slides in HRP-conjugated secondary antibodies (ImmPRESSTM HRP Anti-Rabbit IgG; ImmPRESSTM HRP Anti-mouse IgG) for 30 min.
- Wash off the unbound antibody by a 10 min wash in running tap water.
- Incubate the slides in a 1x working solution of DAB (ImmPACT DAB Peroxidase Substrate SK-4105) for 5 min followed by a 5 min water wash.
- Counter-stain nuclei with Gill's Hematoxylin II for 15 s followed by a water wash and 30 s incubation in Scott’s water followed by a water wash.
- Dehydrate tissues by 1 min sequential immersions in 50% ethanol, 70% ethanol, 95% ethanol, and three times 100% ethanol.
- Incubate the slides in three consecutive containers of xylene for 3 min.
- Mount the slides in DPX (Sigma) according to manufacturer's instructions and leave to dry for 24 h.
- Observe the slides under a microscope (optical) or scan on the Aperio.
- The staining can be scored automatically using the Aperio imaging system.
Pause Point: Once cut and baked, the slides can be stored for a few weeks. Once stained, the slides are stable and can be stored indefinite (see Table 1-[4]).
Caution: It is important not to dry the tissue at any point during the staining process after the rehydration. It is critical always to have a control slide stained only with the secondary antibody (see Table 1-[4]).
- Slice formalin fixed paraffin embedded tissues and then bake at 60 °C for 2 h.
Notes
- Troubleshooting advice can be found in Table 1 below.
Table 1. Troubleshooting advice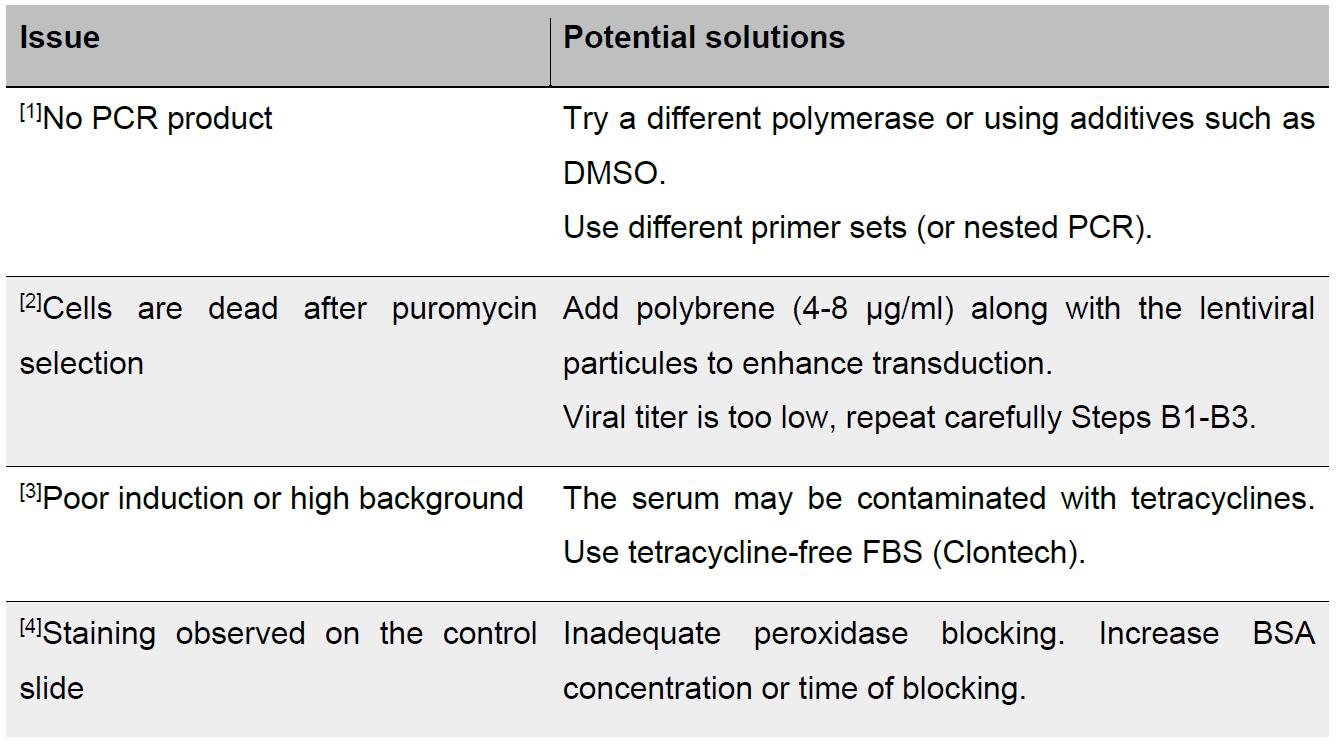
- Examples of ex vivo transduced tissues can be found in McClurg et al. (2018).
Data analysis
Ideally, samples from at least 3 patients (n = 3) should be analyzed and IHC scored independently by 2 trained pathologists.
Recipes
- 2YT media
10 g yeast extract
16 g tryptone
5 g NaCl
Dissolved in 1,000 ml ddH2O and autoclaved - Scott's tap water substitute
2 g/L NaHCO3
20 g/L MgSO4 - Gill's Hematoxylin II solution
4 g/L hematoxylin
Acknowledgments
For this work, OB was supported by the Newcastle’s Biomedical Fellowship Programme, which is in part funded through the Wellcome Trust’s Institutional Strategic Support Fund, and by the Breast Cancer Campaign charity grant number 2013MaySP005. ULM was supported by the JGWP Foundation (BH142412), Newcastle Healthcare Charity (JG/ML/0414), Prostate Cancer UK (PG09-23), and Cancer Research UK (C27826/A15994). KR was supported by the Canadian Institutes of Health Research (CIHR grant MOP-311094).
Competing interests
The authors declare no conflicts of interest or competing interests.
Ethics
As declared in the original publication (McClurg et al., 2018), benign prostatic hyperplasia (BPH) samples were obtained from cancer-free patients as established by the Pathology Department at the Freeman Hospital (Newcastle upon Tyne, UK). The tissue samples were obtained with full ethical approval from Newcastle & North Tyneside 1 NHS Strategic Health Authority Local Research Ethics Committee (reference 15/NE/0400) and patient’s written informed consent.
References
- Cancer Genome Atlas Research, N. (2015). The molecular taxonomy of primary prostate cancer. Cell 163(4): 1011-1025.
- Centenera, M. M., Gillis, J. L., Hanson, A. R., Jindal, S., Taylor, R. A., Risbridger, G. P., Sutherland, P. D., Scher, H. I., Raj, G. V., Knudsen, K. E., Yeadon, T., Australian Prostate Cancer, B., Tilley, W. D. and Butler, L. M. (2012). Evidence for efficacy of new Hsp90 inhibitors revealed by ex vivo culture of human prostate tumors. Clin Cancer Res 18(13): 3562-3570.
- Centenera, M. M., Raj, G. V., Knudsen, K. E., Tilley, W. D. and Butler, L. M. (2013). Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol 10(8): 483-487.
- Feldman, B. J. and Feldman, D. (2001). The development of androgen-independent prostate cancer. Nat Rev Cancer 1(1): 34-45.
- McClurg, U. L., Nabbi, A., Ricordel, C., Korolchuk, S., McCracken, S., Heer, R., Wilson, L., Butler, L. M., Irving-Hooper, B. K., Pedeux, R., Robson, C. N., Riabowol, K. T. and Binda, O. (2018). Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br J Cancer 118(5): 713-726.
- Michael, R. G. and Joseph, S. (2012). Molecular cloning: A laboratory manual. 4th edition. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory Press.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
McClurg, U. L., McCracken, S. R., Butler, L., Riabowol, K. T. and Binda, O. (2018). Ex vivo Culture and Lentiviral Transduction of Benign Prostatic Hyperplasia (BPH) Samples. Bio-protocol 8(21): e3075. DOI: 10.21769/BioProtoc.3075.
Category
Cancer Biology > Oncogenesis > Ex vivo tissue culture model
Cell Biology > Tissue analysis > Tissue culture
Cell Biology > Cell engineering > Lentiviral delivery
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








