- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protocol for in situ Proximity Ligation Assay (PLA) and Microscopy Analysis of Epidermal Growth Factor Receptor (EGFR) Homodimerization
Published: Vol 8, Iss 21, Nov 5, 2018 DOI: 10.21769/BioProtoc.3067 Views: 8823
Reviewed by: Chiara AmbrogioMauro Sbroggio'Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection and Analysis of Circular RNAs by RT-PCR
Amaresh C Panda and Myriam Gorospe
Mar 20, 2018 28793 Views

Microarray, IPA and GSEA Analysis in Mice Models
Stephanie N. Oprescu [...] Shihuan Kuang
Sep 5, 2018 11088 Views

Dual Phospho-CyTOF Workflows for Comparative JAK/STAT Signaling Analysis in Human Cryopreserved PBMCs and Whole Blood
Ilyssa E. Ramos [...] James M. Cherry
Nov 20, 2025 2336 Views
Abstract
Oncogenic drivers play central roles in tumorigenesis as well as in tumor cell survival and proliferation. Mutations of the epidermal growth factor receptor gene (EGFR) that result in constitutive activation of the receptor tyrosine kinase have been identified as oncogenic drivers in a subset of non-small cell lung cancer (NSCLC). PCR-based assays are usually adopted for the detection of EGFR mutations, but no methods to detect EGFR activation that are not based on mutation identification have been established in the clinical setting. We describe a proximity ligation assay (PLA) used to visualize and quantitate EGFR homodimerization in NSCLC cell lines and tissue specimens. Rabbit monoclonal antibodies against EGFR were conjugated to PLUS or MINUS PLA oligonucleotide arms using Probemaker. Annealing of the PLUS and MINUS PLA probes occurred when two EGFR monomers were in close proximity, and repeat sequences in the annealed oligonucleotide complexes were amplified then recognized by a fluorescently-labeled oligonucleotide probe. PLA signals were detected and counted with a fluorescence microscope. We demonstrate the detection of EGFR homodimers by PLA analysis in a quantitative manner in both NSCLC cell lines and tissue samples obtained by transbronchial lung biopsy. PLA methods are a new tool for the detection and quantitation of protein-protein interactions such as homodimers, heterodimers, and fusion proteins.
Keywords: Oncogenic driverBackground
Activating mutations of the epidermal growth factor receptor gene (EGFR) are currently detected by PCR-based assays (Nagai et al., 2005). However, the visual detection and quantitation of activated EGFR in the clinical setting has not been established. In situ proximity ligation assay (PLA) is a technology that uses Duolink® In situ reagents (see References 1 and 2) to create probes by conjugating oligonucleotides to antibodies. When two different types of PLA probes (PLUS and MINUS) are in close proximity (40 nm), annealing occurs which generates an amplified circular DNA. The signal from each detected pair of PLA probes is visualized as an individual spot, and the number of PLA signals per cell can be counted with a fluorescence microscope (Figure 1).
The PLA method can be used to detect any protein-protein interactions in close proximity. We have now applied the PLA using primary antibodies derived from the same species to visualize and quantitate EGFR homodimerization. Furthermore, we detected the formation of EGFR-human epidermal growth factor receptor2 heterodimers and echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase fusion protein in NSCLC cell lines (Ota et al., 2017). This method will be applicable to the detection of other dimerizations and fusions.
Figure 1. In situ PLA principle for EGFR homodimer. A. PLA probes, created by conjugating PLA oligonucleotides and monoclonal antibodies for EGFR, bind to EGFR. B. Connected oligonucleotides hybridize and create multiple circular DNA molecules. C. Fluorescently-labeled detection of oligonucleotides hybridizing to the DNA circle. (Pictures were taken from the Duolink® In situ User Guide [References 1 and 2]).
Materials and Reagents
- Standard pipette tips with a volume capacity of 10 μl, 20 μl, 100 μl, 200 μl, and 1,000 μl (Thermo Fisher Scientific, catalog numbers: 2140, 2149P, ART 10REACH, ART 20P)
- Kimwipe waste paper sheet S-200 (Crecia, catalog number: 62011)
- 12-mm-diameter uncoated cover glasses (Matsunami Glass, catalog number: C012001)
- 24-well plates (Greiner Bio-One, cell culture multiwell plates, catalog number: 66)
- Staining jar (Matsunami, catalog number: No. 13 BT500)
- Dako Pen (Daido Sangyo, PAP-SPAP Pen Super-Liquid Blocker)
- Serological pipettes
- Slide glasses
- NSCLC cell lines: H1975 cells
- Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, Gibco, catalog number: 61870-036)
- Dulbecco's modified Eagle’s medium (DMEM) (Invitrogen, Gibco, catalog number: 12634010)
- Fetal bovine serum (FBS) (Sigma, catalog number: 14C438FBS)
- Penicillin/Streptomycin solution (Invitrogen, catalog number: 15140122)
- Phosphate-buffered saline (PBS) (LSI, catalog number: RM102-PN)
- 4% paraformaldehyde in PBS (Wako, catalog number: 16320145)
- ImmunoSaver (Wako, catalog number: 9706192)
- Duolink® In situ oligonucleotide PLUS (-20 °C) (Sigma-Aldrich, catalog number: DUO92009)
- Duolink® In situ oligonucleotide MINUS (-20 °C) (Sigma-Aldrich, catalog number: DUO92010)
- Conjugation buffer (-20 °C) (Sigma-Aldrich, catalog number: DUO92009)
- Stop reagent (-20 °C) (Sigma-Aldrich, catalog number: DUO92009)
- Storage solution (-20 °C) (Sigma-Aldrich, catalog number: DUO92009)
- 20x assay reagent (-20 °C) (Sigma-Aldrich, catalog number: DUO92009)
- Blocking solution (4 °C) (Sigma-Aldrich, catalog number: DUO92009)
- PLA probe diluent (4 °C) (Sigma-Aldrich, catalog number: DUO92009)
- Rabbit monoclonal antibodies to EGFR (Abcam, catalog number: ab52894)
- 5x Ligation buffer (-20 °C) (Sigma-Aldrich, catalog number: DUO92008)
- 1x Ligase (-20 °C) (Sigma-Aldrich, catalog number: DUO92008)
- 5x Amplification Red (-20 °C) (Sigma-Aldrich, catalog number: DUO92008)
- 1x Polymerase (-20 °C) (Sigma-Aldrich, catalog number: DUO92008)
- Duolink® In situ mounting medium with DAPI (Sigma-Aldrich, catalog number: DUO82040)
- 10% neutral-buffered formalin
- Paraffin
- Xylene
- Ethanol solutions
- Nail polish
- Culture media (10% FBS) (see Recipes)
- Wash buffer A (Sigma-Aldrich, catalog number: DUO82049, see Recipes)
- Wash buffer B (Sigma-Aldrich, catalog number: DUO82049, see Recipes)
Equipment
- Manual pipettes: set of 10 μl, 20 μl, 100 μl, 200 μl, and 1,000 μl (Mettler Toledo, catalog number: Pipet-Lite XLS+ 17014409, 17014412, 17014408, 17014411, 17014407)
- Tweezers
- Freezer
- Direct-Q® 5 UV Remote Water Purification System (Merck, model: Direct-Q® UV 5 Remote, catalog number: ZRQSVR5WW)
- Autoclave (Tomy, catalog number: LSX-500)
- Vortex mixer (Scientific Industries, model: Vortex-Genie 2, catalog number: S1-0286)
- Humidity chamber (Incubation Chamber, Cosmo Bio, catalog number: 10DO)
- Freeze block for enzymes (-20 °C) (Eppendorf, catalog number: 3880001018)
- Fluorescence microscope (Keyence, model: BZ-8100)
Software
- BZ Analyzer software (Keyence)
- Excel (Microsoft)
- GraphPad Prism 5.0 (GraphPad Software)
Procedure
Note: All procedures are performed for four samples.
- Preparation of the cells (Figure 2)
- Place 12-mm-diameter cover glasses in each well of a 24-well plate.
- Grow the cells to 60% confluence on the cover glasses in 500 μl of the recommended culture media at 37 °C, with 5% CO2.
- Remove media from the wells and wash twice each with 1 ml PBS.
- Add 500 μl 4% paraformaldehyde for 15 min at room temperature.
- Remove the solution from the well and wash twice each with 1 ml PBS.
- Take the cover glasses from the wells using tweezers.
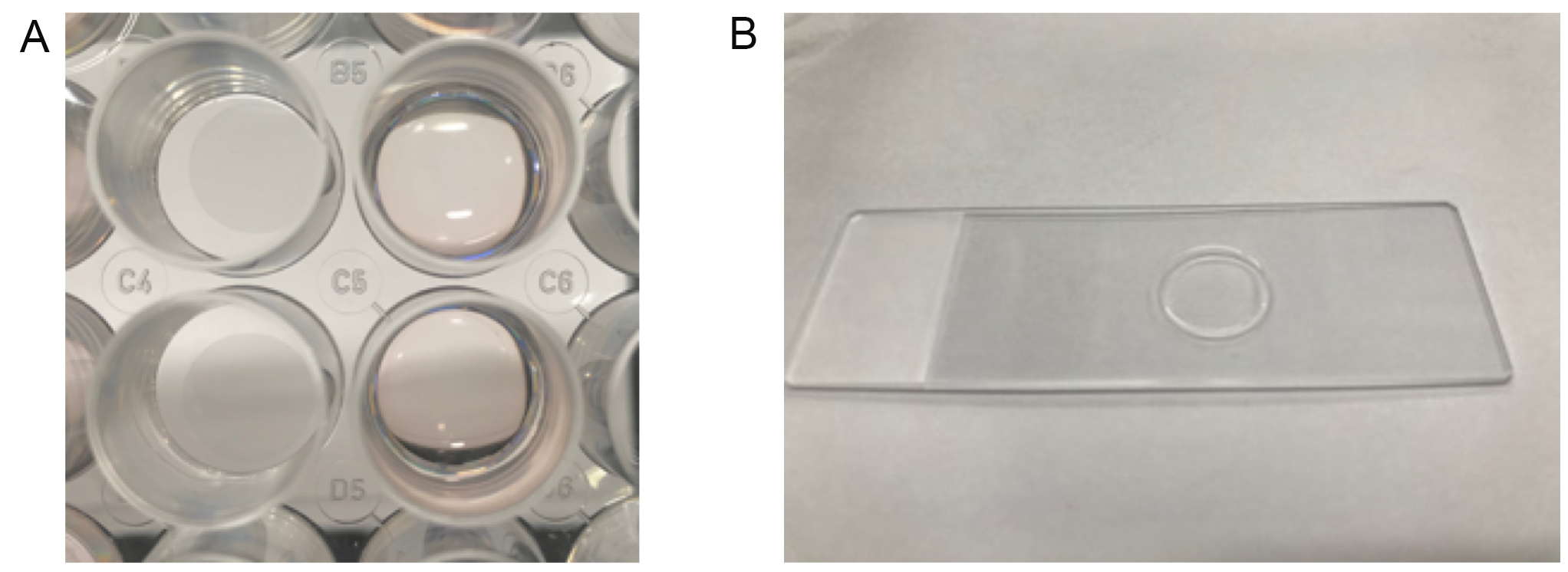
Figure 2. Preparation for PLA method with cell lines. A. Cover glasses in each well of the plate (left). Cells grown on cover glasses in the culture media (right). B. Post-PLA slide glasses mounted with cover glasses using DAPI and sealed with nail polish. - Preparation of the tumor specimens
- Fix surgical specimens in 10% neutral-buffered formalin, embed them in paraffin, and section at a thickness of 4 μm.
- Remove paraffin with xylene, rehydrate with a graded series of ethanol solutions, and wash twice with PBS.
- Add 300 μl ImmunoSaver to 60 ml Milli-Q water in the staining jar.
- Autoclave the slides in the staining jar for 45 min at 98 °C and incubate at room temperature.
- Wash the slides twice with PBS.
- Draw a circle around the tumor specimen of around 1 cm2 with the Dako Pen.
- Conjugation of the PLA oligonucleotide to the antibody
- Thaw conjugation buffer at room temperature and vortex.
- Add 2.1 μl conjugation buffer to 21 μl rabbit monoclonal anti-EGFR antibody and mix gently with a pipette.
- Transfer the antibody solution from Step C2 to one vial of oligonucleotide PLUS or oligonucleotide MINUS and mix gently with a pipette.
- Incubate at room temperature overnight.
- Vortex stop reagent solution and add 2.1 μl to the solution from Step C3 and incubate at room temperature for 30 min.
- Vortex storage solution and add 25.2 μl to the solution prepared in Step C5 and store at 4 °C.
- The stored solution can be used within one month.
- Blocking
- Vortex blocking solution and transfer 40 μl to each sample.
- Put the slides in a pre-heated humidity chamber and incubate in a humidified 37 °C, 5% CO2 incubator for 30 min.
- PLA probes
- Dilute the conjugated antibodies from Step C6 in the PLA probe diluent.
- Prepare 200 μl of 1x working assay reagent by mixing 10 μl of 20x stock solution and 190 μl of PLA probe diluent.
- Add 8 μl PLUS or MINUS antibody solution (Step C6) to 72 μl working assay reagent (from Step E1a) and incubate for 20 min at room temperature.
- Gently remove the blocking solution from the samples using a Kimwipe waste paper sheet without touching the cells.
- Add 38 μl (19 μl PLUS probe and 19 μl MINUS probe) in total diluted PLA probes PLUS solution and MINUS solution to each sample.
- Mix gently with a pipette.
- Incubate in a humidity chamber for 2 h at room temperature.
- Dilute the conjugated antibodies from Step C6 in the PLA probe diluent.
- Ligation
- Dilute 32 μl ligation buffer (5x) in 124 μl Milli-Q water and mix.
- Gently remove the PLA probe solution from the samples using a Kimwipe waste paper sheet without touching the cells.
- Gently wash the slides twice for 5 min each in 500 μl Wash Buffer A.
- Add 4 μl ligase to 156 μl ligation buffer (1x) and vortex.
- Add 39 μl solution to each sample.
- Put the slides in a pre-heated humidity chamber and incubate in a humidified 37 °C, 5% CO2 incubator for 30 min.
- Amplification
- Dilute 32 μl amplification red (5x) in 126 μl Milli-Q water and mix.
- Gently remove the ligation solution from the samples using a Kimwipe waste paper sheet without touching the cells.
- Gently wash the slides twice for 2 min each in 500 μl Wash Buffer A.
- Add 2 μl polymerase solution to 158 μl amplification red (1x) and vortex.
- Add 39 μl solution to each sample.
- Put the slides in a pre-heated humidity chamber and incubate with a humidified 37 °C, 5% CO2 incubator for 30 min.
- Preparation for imaging
- Gently remove the amplification-polymerase solution from the samples using a Kimwipe waste paper sheet without touching the cells.
- Gently wash the slides twice for 10 min each in 500 μl Wash Buffer B.
- Gently wash the slides for 1 min in 500 μl 0.01x Wash Buffer B.
- Let the slides dry at room temperature in the dark.
- Mount the slide glasses with cover glasses using a minimal volume of Duolink® In situ Mounting Medium with DAPI.
- Seal the edges with nail polish and incubate for 15 min at room temperature in the dark.
- Imaging with a microscope
- Analyze the images using fluorescence microscopy.
- After imaging, store the slides at -20 °C in the dark.
Data analysis
- PLA signals are detected with a Keyence BZ-8100 fluorescence microscope (Figure 3).
- PLA signals are visible as red spots, and are detected in NSCLC cells in a manner dependent on the addition of both PLUS and MINUS probes, indicating the presence of EGFR homodimers.
- Nuclei are stained blue with 4’,6-diamidino-2-phenylindole (DAPI).
- The number of PLA signals per cell is quantified by counting the number of red spots in 20 cells using BZ Analyzer software.
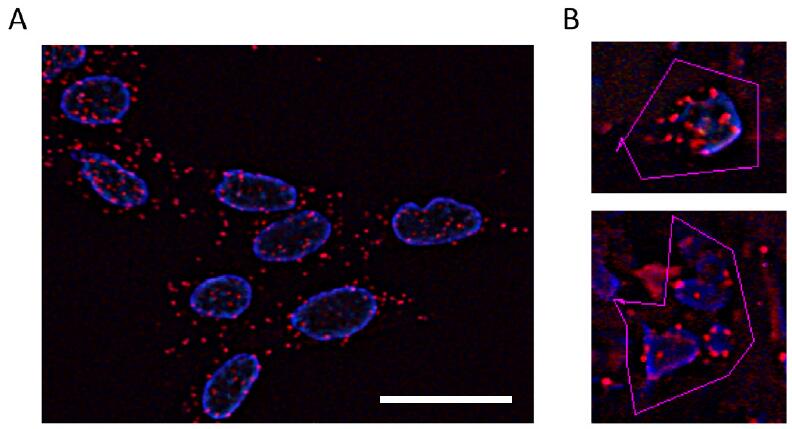
Figure 3. Detection of EGFR homodimerizations in NSCLC cell lines and resected specimens by PLA. Representative images of EGFR homodimers in H1975 cells (A) and in lung adenocarcinoma surgical specimens (B). The positive and negative controls are provided in Ota et al. (2017). Tumor cells surrounded by pink lines to count the number of red spots per cell by analytical software (B). Scale bar = 20 μm.
Notes
- The concentration of the primary antibodies before adding the conjugation buffer is desirably more than 1 mg/ml, and should at least be higher than that used for immunohistochemistry.
- Antibodies derived from any animal are permitted, we use rabbit antibodies.
- All experiments with detection reagents (from Procedure G onwards) are performed in a dark room.
Recipes
- Culture media (10% FBS)
- Open liquid DMEM or RPMI bottle (500 ml)
- Take out 55 ml with a sterile serological pipette and discard the liquid
- Add 50 ml FBS with a sterile serological pipette
- Add 5 ml Penicillin/Streptomycin solution
- Store at 4 °C
- Wash buffer A
Dissolve the contents of one pouch in pure water to a final volume of 1,000 ml and store at 4 °C - Wash buffer B
Dissolve the contents of one pouch in pure water to a final volume of 1,000 ml and store at 4 °C
Acknowledgments
We thank Akiko Sato for technical assistance. We thank Sarah Williams, PhD, from Edanz Group (www.edanzediting.com) for editing a draft of this manuscript. This work was supported in part by JSPS KAKENHI Grant Number JP18K15927.
Competing interests
No potential conflicts of interest were disclosed.
References
- Duolink® In situ - Fluorescence User Guide. Sigma-Aldrich. (Accessed 4-July, 2018, at https://reedd.people.uic.edu/ReedLabPLA.pdf.)
- Duolink® In situ - Probemaker User Guide. Sigma-Aldrich. (Accessed 4-July, 2018, at https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma-Aldrich/Instructions/1/duolink-probemaker-user-manual.pdf.)
- Nagai, Y., Miyazawa, H., Huqun, Tanaka, T., Udagawa, K., Kato, M., Fukuyama, S., Yokote, A., Kobayashi, K., Kanazawa, M. and Hagiwara, K. (2005). Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res 65(16): 7276-7282.
- Ota, K., Harada, T., Otsubo, K., Fujii, A., Tsuchiya, Y., Tanaka, K., Okamoto, I. and Nakanishi, Y. (2017). Visualization and quantitation of epidermal growth factor receptor homodimerization and activation with a proximity ligation assay. Oncotarget 8(42): 72127-72132.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ota, K. and Harada, T. (2018). Protocol for in situ Proximity Ligation Assay (PLA) and Microscopy Analysis of Epidermal Growth Factor Receptor (EGFR) Homodimerization. Bio-protocol 8(21): e3067. DOI: 10.21769/BioProtoc.3067.
Category
Cancer Biology > General technique > Molecular biology technique
Molecular Biology > Protein > Phosphorylation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link