- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Innovative Approach to Study Ralstonia solanacearum Pathogenicity in 6 to 7 Days Old Tomato Seedlings by Root Dip Inoculation
Published: Vol 8, Iss 21, Nov 5, 2018 DOI: 10.21769/BioProtoc.3065 Views: 7533
Reviewed by: Samik BhattacharyaMahmoud Kamal AhmadiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1342 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 404 Views
Abstract
Ralstonia solanacearum (F1C1) is a Gram-negative plant pathogenic bacterium that causes lethal wilt disease in a wide range of plant species. This pathogen is very well known for its unpredictable behavior during infection and wilting its host. Because of its mysterious infection behavior, virulence and pathogenicity standardization are still a big challenge in the case of R. solanacearum. Here, we report an innovative pathogenicity assay of R. solanacearum (F1C1) in the early stage of tomato seedlings by root dip inoculation. In this assay, we employed 6-7days old tomato seedlings for infection grown under nutrients free and gnotobiotic condition. After that, pathogenicity assay was performed by maintaining the inoculated seedlings in 1.5 or 2 ml sterile microfuge tubes. During infection, wilting symptom starts appearing from ~48 h post inoculation and the pathogenicity assay gets completed within seven days of post inoculation. This method is rapid, consistent as well as less resource dependent in terms of labor, space and cost to screen large numbers of plants. Hence, this newly developed assay is an easy and useful approach to study pathogen virulence functions and its interaction with the host plant during wilting and disease progression at the seedling stage.
Keywords: Ralstonia solanacearumBackground
Bacterial wilt pathogen Ralstonia solanacearum dwells in soil. When favorable condition comes, the bacterium enters inside the suitable host plant through root, grows, colonizes there and ultimately kills the plant. R. solanacearum strain (F1C1) had been isolated and characterized from a wilted chilli plant nearby Tezpur (Assam), north-east India (Kumar et al., 2013). Owing to the lethality and its exceptional wide host range, R. solanacearum considered as the second most devastating bacterial phytopathogen around the world (Mansfield et al., 2012). In spite of that, no effective approach so far is available to deal with this pathogen and its associated disease.
Regarding its virulence and pathogenicity assay, several existing methods are commonly in use such as soil drenching, leaf clip, petiole cut as well as stem inoculation but, in the other hand it is also reported that these methods are not found to be very much appropriate in analyzing minute virulence and pathogenicity differences in few mutants strain of R. solanacearum (Macho et al., 2010). In the wake of developing a potent pathogenicity assay, we have developed and standardized an innovative root inoculation method to study R. solanacearum pathogenicity in tomato seedling (Singh et al., 2018). Along with the pathogenicity assay (Figure 1), this approach is also equally helpful in bacterial bio-control development and plant protection assay against this wilt pathogen at the seedling stage. Recently many other research groups also have employed seedling stages of tomato plants for studying R. solanacearum pathogenicity in number of occasions (Pradhanang et al., 2000; Artal et al., 2012; Kumar, 2014; Kumar et al., 2017). This vascular pathogen is well known for its mysterious infection behavior and wide range of host adaptability, therefore R. solanacearum is also referred as a suitable model to investigate fundamental aspects of plant-pathogen interaction and host adaptations (Genin and Boucher, 2002; Genin, 2010; CollandValls, 2013; Singh et al., 2018).
Materials and Reagents
- 1.5 ml and 2.0 ml micro centrifuge tubes (Tarson, catalog numbers: 500010 and 500020)
- Pipette tips (10 μl) (Tarson, catalog number: 521000)
- Pipette tips (200 μl) (Tarson, catalog number: 521010)
- Pipette tips (1,000 μl) (Tarson, catalog number: 521020)
- 13 mm screw vial (Borosil, catalog number: VO04C113005000)
- 30 ml culture tubes (Riviera, catalog number: 71200305)
- 50 ml Falcon tube (Tarson, catalog number: 546041)
- 250 ml conical flask (Borosil, catalog number: 4980021)
- Plastic tray (13 cm x 22 cm)
- Petri dishes (90 mm) (Tarson, catalog number: 460090)
- 120-well 1.5 ml to 2.0 ml tube rack (Tarson)
- Meta loop (Himedia, catalog number: LA650-1)
- Cotton wool(SN surgical & health care science (P) LTD, catalog number: DL-160M)
- Tissue paper (Hygienics Plus, catalog number: HPTP02)
- Ralstonia solanacearum (F1C1) (Lab collection)
- Tomato seeds (Durga: Selection-22) [Durga seed farm(REGD), Chandigarh, India]
- Sterile distilled water
- Peptone (Himedia, catalog number: RM001-500G)
- Casamino acid or casein acid hydrolysate (SRL, catalog number: CI019)
- Glucose (Himedia, catalog number: MB037-500G)
- Agar powder (Himedia, catalog number: GMR026-500G)
- Yeast extract powder (Himedia, catalog number: RM027-500G)
- BG agar media (see Recipes)
- BG broth media (see Recipes)
- 20% Glucose supplement (see Recipes)
Equipment
- Pipettes (Eppendorf-these pipettes can accommodate pipette tips of 10 μl, 200 μl, 1,000 μl respectively)
- Water distillation system (Riveria, catalog number: 7222010)
- Autoclave (EquitronMedica, model: 7431PAD)
- Incubator and shaker (28 °C) (Scigenics Biotech, Orbitek®, model: LE0102DCBA)
- Macro centrifuge (Eppendrof, model: 5804R)
- Laminar Air Flow (Ikon Instruments, model: 1K-137)
- Weighing balance (Mettle Toledo, model: ME204)
- Growth chamber (ScigenicsBiotech, Orbitech®, model: GC350)
Procedure
- Germination of tomato seedling (Figure 1)
- Take tomato seeds and wash them with sterile distilled water.
- Soak seeds in sterile distilled water for 48 h at room temperature of ~25 to 30 °C (Figure 1A).
- Spread soaked seeds on cotton and tissue bed in a plastic or glass tray, and then put the tray for germination in a growth chamber maintained at 28 °C, 75% RH and 12 h of photoperiod (Light intensity of the lamp as 1,350 lm and the color temperature as 4,000 K) (Figure 1B).
- Maintain the moisture of the seed bed by spraying sterile distilled water at regular interval during the seed germination.
Note: The spray of sterile distilled water at regular interval help in better seedling growth and it also helps to prevent bacterial and/or fungal contamination. - Pick the 6-7 days old tomato seedlings with similar size for infection assay.
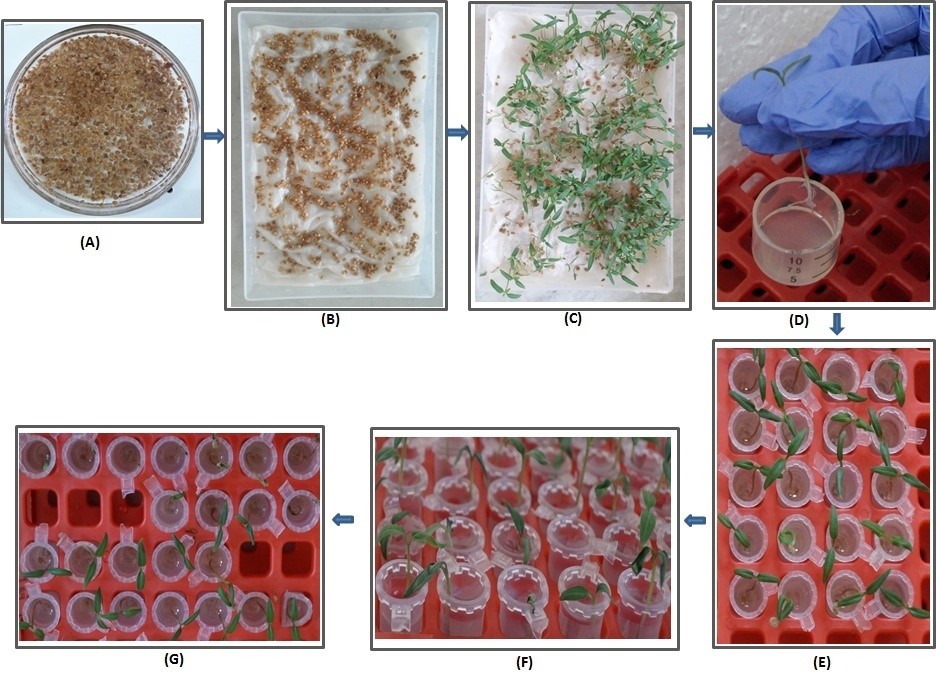
Figure 1. Pictures showing the different steps of root inoculation assay to study R. solanacearum pathogenicity in tomato seedlings. A. Soaking of tomato seeds in sterile distilled water for 48 h. B. Spreading of seeds on sterile and wet cotton and tissue paper bed. C. Germination of tomato seeds on a sterile and wet tissue paper bed in a growth chamber at 28 °C and 75% relative humidity up to the seedling stage. D. Root inoculation of tomato seedling by dipping the root in bacterium inoculums container. E. Each inoculated seedling transferred to sterile1.5-2 ml empty microfuge tubes. After approximately 5 min of air exposure, 1.0 to 1.5 ml of sterile water was added to each microfuge tube. F. Approximately 40-48 h post inoculation, infected seedlings started wilting indicating the early wilting symptom. G. On the 7th day post inoculation, 80% to 90% of infected tomato seedlings became permanently wilted and died (top side) in comparison to water as control (bottom side). - Bacterial inoculum preparation
- Streak R. solanacearum (F1C1) bacterium on BG-Agar plate and incubate in an incubator (Orbitek, Scigenics Biotech (Pvt.) Ltd., India) maintained at 28 °C for 24 h.
- Add freshly grown R. solanacearum (F1C1) colonies to 50 ml BG broth in a sterile vial with a sterile loop and allow to grow in a shaking incubator (Orbitek, Scigenics Biotech (Pvt.) Ltd., India) maintained at 28 °C and 150 rpm for 24 h.
- Centrifuge the full-grown bacterial cultures at 4,000 rpm (3,155 x g) for 15 min at 4 °C and then discard the supernatant.
- Resuspend bacterial pellets in sterile distilled water and repeat Step B3 (centrifuge at 4,000 rpm [3,155 x g] for 15 min at 4 °C) to remove remaining culture media component from the suspension.
- Now resuspend bacterial pellet in an equal volume of sterile distilled water to obtain a concentration of ~109 CFU/ml. Now, this bacterial inoculum is ready for infection.
- Inoculation of R. solanacearum in 6-7 days old tomato seedlings (Figure 2)
- Pick 6-7 days old tomato seedlings with similar size from germination bed.
- Add 5-20 ml of bacterial inoculum into a wide mouth container.
- Immerse the seedling roots in bacterial inoculum up to the root-shoot junction for a few seconds.
- Now transfer these seedlings to an empty and sterile microfuge (1.5 or 2.0 ml) tube.
- Then air expose the seedlings for ~5 min (Here “air expose” means that pathogen inoculated seedlings are placed in a sterile empty microfuge tube for ~5 min before addition of water to the tube).
- Add 1 to 1.5 ml of sterile distilled water to the microfuge tube containing the air exposed seedlings.
- Inoculate seedlings in the same way with sterile distilled water as a negative control.
- Transfer all the inoculated as well as control seedlings to a growth chamber maintained at 28 °C, 75% RH, and 12 h of photoperiod.
- Start infection and disease progression analysis from the next day onwards till the 7th day post inoculation.
Figure 2. Schematic diagram of root inoculation of tomato seedlings in the R. solanacearum pathogenicity assay. Here, we are representing self-explanatory schematic diagram of pathogenicity assay for easy and better understanding of the crucial steps of the assay.
Data analysis
In this work, we have successfully described details of an easy and efficient approach to study R. solanacearum pathogenicity in the early seedlings stage of tomato cultivars. The described assay is very much reproducible and appropriate to analyze the different kind of mutant gene function including minute virulence function in R. solanacearum.
In addition to above, during infection assay, we have observed and first time reported an interesting air influence (Air exposed pathogen inoculated tomato seedling shown more aggressive and consistent disease progression in comparison to unexposed one) phenomenon on R. solanacearum pathogenicity in tomato seedlings by root inoculation. After bacterial inoculation, tomato seedlings will appear wilting symptom within 48 h and became completely wilted within 7 days post inoculation. This inoculation method mimics natural mode of infection caused by this wilt pathogen. Considering the reproducibility and efficiency of this root inculcation method in early seedling stage of tomato, it would help the concerned scientific community to study the virulence behavior, host adaptability as well as many other fundamental aspects of plant-microbe interaction during R. solanacearum infection and disease progression.
All the other required information about data processing, statistical analysis as well as details of replicates and independent experiments were already included in original research paper Singh et al. (2018).
Notes
- As tomato seeds germination are not 100% efficient, this should be taken into consideration while designing the experiment.
- Add some amount of water to the germination bed just before starting the tomato seedling picking for root inoculation which will help in smooth picking as well as minimizing the injury or breakage in root seedling.
- Although we have maintained ~5 min of air exposure to get better infection result but, postinoculation air exposure for a fraction of second is also enough to induce disease symptom.
- This assay was validated with the help of non-pathogenic bacterium strain inoculation as well as different mutant derivative strains of R. solanacearum (F1C1) such as hrpB, phc A and pilT. After that, further bacterial colonization studies inside inoculated and wilted seedlings were also confirmed by GUS staining and mCherry-fluorescence (Singh et al., 2018).
- All these bacterial inoculation and infection steps were performed on clean and surface sterilized workbench inside the laboratory. For infection and pathogenicity assay steps sterile hood with laminar flow would not be required.
Recipes
- BG-Agar, pH 6.5-7 (100 ml)
1 g Peptone
0.1 g Casamino acid
0.1 g Yeast extract
1.5 g Agar
Add distilled water up to 100 ml and autoclave the media at 121 °C (249 °F) for around 15-20 min
Supplement 500 μl of 20% glucose to it and pour into sterile Petri plates, let the plates completely solidify - BG-Broth, pH 6.5-7 (100 ml)
1 g Peptone
0.1 g Casamino acid
0.1 g Yeast extract
Add distilled water up to 100 ml and autoclave the media at 121 °C (249 °F) for around 15-20 min. Supplement 500 μl of 20% glucose to it - 20% Glucose supplement (5 ml)
1 g glucose
Add distilled water up to 5 ml and autoclave it at 121 °C (249 °F) for around 15-20 min
Acknowledgments
Niraj Singh is very much thankful to DBT, Government of India for the fellowship (DBT-JRF & SRF). Rahul Kumar is thankful to the CEFIPRA and CSIR, Govt. of India for the post doctoral fellowships. Suvendra Kumar Ray is grateful to CEFIPRA for the Indo-French project grant (4800-B1). Apart from these, S. K. Ray’s laboratory research is also supported by various departmental projects such as UGC-SAP (DSR II), DST-FIST, and DBT-Strengthening NE. We are also thankful to S. Genin, LIPM of France, L. Saho of IIT-Guwahati, India, as well as lab members Anjan Barman, Tarinee Phukan, Pankaj Losan Sharma, Ruksana Aziz and Kristi Kabyashree for their coordination and kind support.
Competing interests
The authors have declared no conflict of interest.
References
- Artal, R. B., Gopalkrishnan, C. and Thippeswamy, B. (2012). An efficient inoculation method to screen tomato, brinjal and chilli entries for bacterial wilt resistance. Pest Manage Hortic Ecosyst 18(1): 70-73.
- Coll, N. S. and Valls, M. (2013). Current knowledge on the Ralstonia solanacearum type III secretion system. Microb Biotechnol 6(6): 614-620.
- Genin, S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstoniasolanacearum. New Phytol 187(4): 920-928.
- Genin, S. and Boucher, C. (2002). Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Mol Plant Pathol 3(3): 111-118.
- Kumar, R. (2014). Studying virulence functions of Ralstonia solanacearum, the causal agent of bacterial wilt in plants. Ph.D. thesis. Tezpur University, Tezpur, India.
- Kumar, R., Barman, A., Jha, G. and Ray, S. K. (2013). Identification and establishment of genomic identity of Ralstonia solanacearum isolated from a wilted chilli plant at Tezpur, North East India. Curr Sci 105(11): 1571-1578.
- Kumar, R., Barman, A., Phukan, T., Kabyashree, K., Singh, N., Jha, G., Sonti, R. V., Genin, S. and Ray, S. K. (2017). Ralstonia solanacearum virulence in tomato seedlings inoculated by leaf clipping. Plant Pathol 66(5): 835-841.
- Macho, A. P., Guidot, A., Barberis, P., Beuzon, C. R. and Genin, S. (2010). A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol Plant Microbe Interact 23(9): 1197-1205.
- Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., Dow, M., Verdier, V., Beer, S. V., Machado, M. A., Toth, I., Salmond, G. and Foster, G. D. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13(6): 614-629.
- Pradhanang, P. M., Elphinstone, J. G. and Fox, R. T. V. (2000). Identification of crop and weed hosts of Ralstonia solanacearum biovar 2 in the hills of Nepal. Plant Pathol 49(4): 403-413.
- Singh, N., Phukan, T., Sharma, P. L., Kabyashree, K., Barman, A., Kumar, R., Sonti, R. V., Genin, S. and Ray, S. K. (2018). An innovative root inoculation method to study Ralstonia solanacearum pathogenicity in tomato seedlings. Phytopathology 108(4): 436-442.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Singh, N., Kumar, R. and Ray, S. K. (2018). An Innovative Approach to Study Ralstonia solanacearum Pathogenicity in 6 to 7 Days Old Tomato Seedlings by Root Dip Inoculation. Bio-protocol 8(21): e3065. DOI: 10.21769/BioProtoc.3065.
Category
Microbiology > Microbe-host interactions > Bacterium
Plant Science > Plant immunity > Host-microbe interactions
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











