- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
H1N1 Virus Production and Infection
Published: Vol 8, Iss 20, Oct 20, 2018 DOI: 10.21769/BioProtoc.3062 Views: 7388
Reviewed by: Alka MehraMigla MiskinyteAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Participant-Derived Xenograft Mouse Model to Decode Autologous Mechanisms of HIV Control and Evaluate Immunotherapies
Emma Falling Iversen [...] R. Brad Jones
Apr 5, 2025 2517 Views

Analysis of Vascular Permeability by a Modified Miles Assay
Hilda Vargas-Robles [...] Michael Schnoor
Apr 5, 2025 2543 Views

PBMC-Humanized Mouse Model for Multiple Sclerosis: Studying Immune Changes and CNS Involvement
Anastasia Dagkonaki [...] Lesley Probert
May 20, 2025 3977 Views
Abstract
Influenza A virus is a member of orthomyxoviridae family causing wide-spread infections in human respiratory tract. Mouse infection model is widely used in antiviral research and pathogenesis study against influenza A virus. Here, we report a protocol in infected mice with different virus doses and strains to explore how an inhibitor of lysine-specific demethylase (LSD1) impacts disease progression.
Keywords: Influenza A virusBackground
Influenza A virus, a member of the family Orthomyxoviridae, is a negative-sense RNA virus with eight segmented genomes of single-stranded viral RNAs (vRNAs) that encode more than 10 proteins. During the past 100 years, outbreaks of influenza-virus strains regularly appeared in human populations, including “Spanish flu” in 1918 caused by the H1N1 subtype, “Asian flu” in 1957 by H2N2, “Hong Kong flu” in 1968 by H3N2, “Russian flu” in 1977 by H1N1, and “swine flu” in 2009 by H1N1 (Smith et al., 2009; Lim and Mahmood, 2011; Kumar et al., 2018). Seasonal influenza A viruses also circulate worldwide, spread easily from person to person, and result in the hospitalization of three to five million individuals worldwide annually (Molinari et al., 2007). The seasonal influenza infections are responsible for 290,000-650,000 deaths annually, mainly among young children, elderly adults, and critically ill patients (Kumar et al., 2018).
Animal models are used in influenza virus research not only to elucidate the viral and host factors that affect disease outcomes and spread among susceptible hosts but also to evaluate interventions designed to prevent or reduce influenza morbidity and mortality (Thangavel and Bouvier, 2014). In this paper, we use two strains of influenza A virus, A/WSN/33(H1N1) (WSN) which is a commonly-used lab strain, and A/Sichuan/01/2009 (SC09) which is a natural isolate to infect mice. This experiment aims to explore how Trans-2-phenylcyclopropylamine hydrochloride (TCP) (a chemical inhibitor against LSD1 [Shan et al., 2017]) impacts disease progress. Moreover, we applied different doses of virus to infect the mice for different purposes to reveal the function of TCP.
Materials and Reagents
- 1.5 ml Eppendorf tubes (Eppendorf)
- 10 µl, 200 µl, and 1 ml pipette tips (FUKAEKASEI and WATSON, catalog number: 1201-705C )
- 24-well plates (Corning, catalog number: 3526 )
- 6-well plates(Corning, catalog number: 353046 )
- 6 cm dish (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150288 )
- Syringes
- 75 cm flask (T75 flask) (Corning, catalog number: 430641U )
- 0.22 μm filters (Axiva Sichem Biotech, catalog number: SFPV13 R )
- Pencil
- BALB/c mice (6-8W, female)
- 293T cells (ATCC, catalog number: CRL-3216 )
- MDCK cells (ATCC, catalog number: CCL-34 )
- A/WSN/33(H1N1) (WSN) virus (Hoffmann et al., 2000)
- A/Sichuan/1/2009 (H1N1) (SC09) (Kindly provided by Prof. Yuelong Shu in China CDC)
- TCP (Santa Cruz Biotechnology, catalog number: sc-208452 )
- 1x PBS (Lonza, catalog number: 17-516Q )
- Isoflurane (Abbott, catalog number: 5260-04-05)
- Picric acid (Fisher Scientific, catalog number: 13205 ) (Used for labeling the mice)
- DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: 11965092 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- True Blue substrate (KPL, catalog number: 50-78-02 )
- anti-nucleoprotein antibody (Antibody Research Centre, Shanghai Institute of Biological Science)
- Anti-rabbit IgG secondary antibody (Antibody Research Centre, Shanghai Institute of Biological Science)
- Avicel (FMC BioPolymer, catalog number: CL 611 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912 )
- 2x DMEM (Thermo Fisher Scientific, catalog number: 12800017 )
- Trypsin (TPCK) (Sigma-Aldrich, catalog number: 4370285 )
- Fetal bovine serum heat inactivated (FBS) (Sigma-Aldrich, catalog number: F9665 )
- BactoTM agar (BD, BactoTM, catalog number: 214010 )
- 18 MΩ H2O
- Overlay medium (DMEM + 2% FBS + 0.9% BactoTM-Agar)
- Infection medium (DMEM + 1 μg/ml TPCK)
- Trans-2-phenylcyclopropylamine hydrochloride (TCP) (1 mg/ml) (see Recipes)
- 2.4% Avicel (see Recipes)
- 6% BSA (see Recipes)
- Overlay medium (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, catalog number: 1156-6963 )
- Dissection equipment (forceps, tweezers, scissors)
- Anesthesia machine (Parkland Scientific, catalog number: V3000PK )
- Eppendorf centrifuge
- Class II biological safety hood (Thermo Fisher Scientific)
- Incubator
- Freezer (4 °C, -20 °C and -80 °C)
- Ultraviolet (UV) light
Software
- GraphPad Prism (https://www.graphpad.com/)
- Statistical Product and Service Solutions (SPSS) (https://www.ibm.com/analytics/data-science/predictive-analytics/spss-statistical-software)
Procedure
- Virus production
Generate WSN virus by reverse genetics as described previously in biosafety level 2 hood (Hoffmann et al., 2000). A/Sichuan/1/2009 [H1N1] [SC09] virus was kindly provided by Prof. Yuelong Shu in China CDC.- Transfection and transduction
- Transfect monolayer of 293T cells in a 6 cm dish with 1 μg each of eight plasmids of pHW2000-WSN-segment1-8 (kindly provided by Prof. Hans Klenk, Germany) by following the manufacturer’s protocol.
- Twenty-four hours post-transfection, change the medium to 4 ml DMEM with 2% FBS, and culture at 37 °C for another 48 h.
- Transfer all the supernatant from the transfected 293T cells to the monolayer of MDCK cells in the 6 cm dish, fill with fresh DMEM with 2% FBS if the whole volume is less than 4 ml.
- Culture the MDCK cells for 48-96 h till CPE can be observed. Collect the MDCK supernatant and make a stock.
- Plaque purification
- Dilute the stock supernatant in serial 10-times dilution in PBS and apply 300 μl of each dilution onto the indicated wells of monolayer of MDCK cells plated in 6-well plate for 0.5 h.
- After this, discard the residual medium and wash the cells in PBS for 2 times. Then add 2 ml of overlay medium (DMEM + 2% FBS + 0.9% BactoTM-Agar) on the cells and culture the cells for 2-3 days to observe plaques.
- Encircle the plaque from beneath with a pencil and pick one plaque from the top with a 1 ml-pipette-tip, and then resuspend the plaque in 1 ml PBS at 4 °C for 16 h. This 1 ml PBS containing viruses will be used as the first-round plaque to repeat plaque purification for a second time.
- Sequence the resulted virus and multiply in MDCK cells in a 75 cm flask to make a virus stock.
- Transfection and transduction
- Virus amplification and virus titer determinations
- Virus amplification
- Seed monolayers of MDCK cells in a T75 flask, after 12 h cells should be 80% confluent. Then infect cells with virus at MOI = 0.001 in 15 ml infection medium (DMEM + 1 μg/ml TPCK).
- Collect virus supernatant when cells are almost dead (about 3 days). Centrifuge at 1,000 x g for 10 min, sub-pack the supernatant and keep them in -80 °C.
- Determine viral titer by plaque assay (Matrosovich et al., 2016)
- Seed monolayers of MDCK cells in 24-well plates, after 12 h cells should be 100% confluent.
- Then infect cells with 200 µl virus supernatants which are in ten-fold serial dilution by DMEM.
- After a 1 h incubation at 37 °C in 5% CO2, remove the medium and wash once with 1x PBS.
- Then add 2 ml of overlay medium to cover the MDCK cells and incubate for 2 days, subsequently fix the cells in 4% paraformaldehyde and exposure to ultraviolet (UV) light for 30 min.
- Perform immunostaining using an anti-nucleoprotein polyclonal primary antibody for 1 h at room temperature and an HRP-conjugated anti-rabbit IgG secondary antibody for 2 h at room temperature.
- Finally, add 100 µl True Blue substrate to visualize the plaques.
- Virus amplification
- The measurement of median lethal dose
- Transfer the mice (BALB/c mice aged 6-8 weeks) to ABSL2 for adaptive feeding for 3-7 days. Divide the mice (BALB/c mice aged 6-8 weeks) randomly into four groups to infect different concentrations of influenza A virus, 4 mice for each group.
- Dilute virus in 50 μl PBS at different concentrations (4 x 104 pfu/mouse, 4 x 103 pfu/mouse, 4 x 102 pfu/mouse, 4 x 101 pfu/mouse) for WSN and ( 9 x 105 pfu/mouse, 1.3 x 105 pfu/mouse, 1.3 x 104 pfu/mouse, 1.3 x 103 pfu/mouse ) for SC09 (H1N1) strains.
- Anesthetize the mice with isoflurane and infect the mice intranasally by 50 μl droplets containing different concentrations of viruses diluted in PBS.
- Monitor the weight and death of the mice throughout the infection time course from Day 0 to Day 14 (Figure 1).
- Calculate the median lethal dose by Statistical Product and Service Solutions (SPSS)
To run the probit analysis in SPSS, we follow the steps as follow: Firstly, input a minimum of three columns into the Data Editor (Number of individuals per container that responded, Total of individuals per container, Concentrations). Secondly, after columns are set, go to analyze, and press regression, then press probit. Thirdly, set your number responded column as the “Response Frequency”, the total number per container as the “Total Observed”, and the concentrations as the “Covariates”. Don’t forget to select the log base 10 to transform your concentrations. Press “OK” and then get the LD50 results.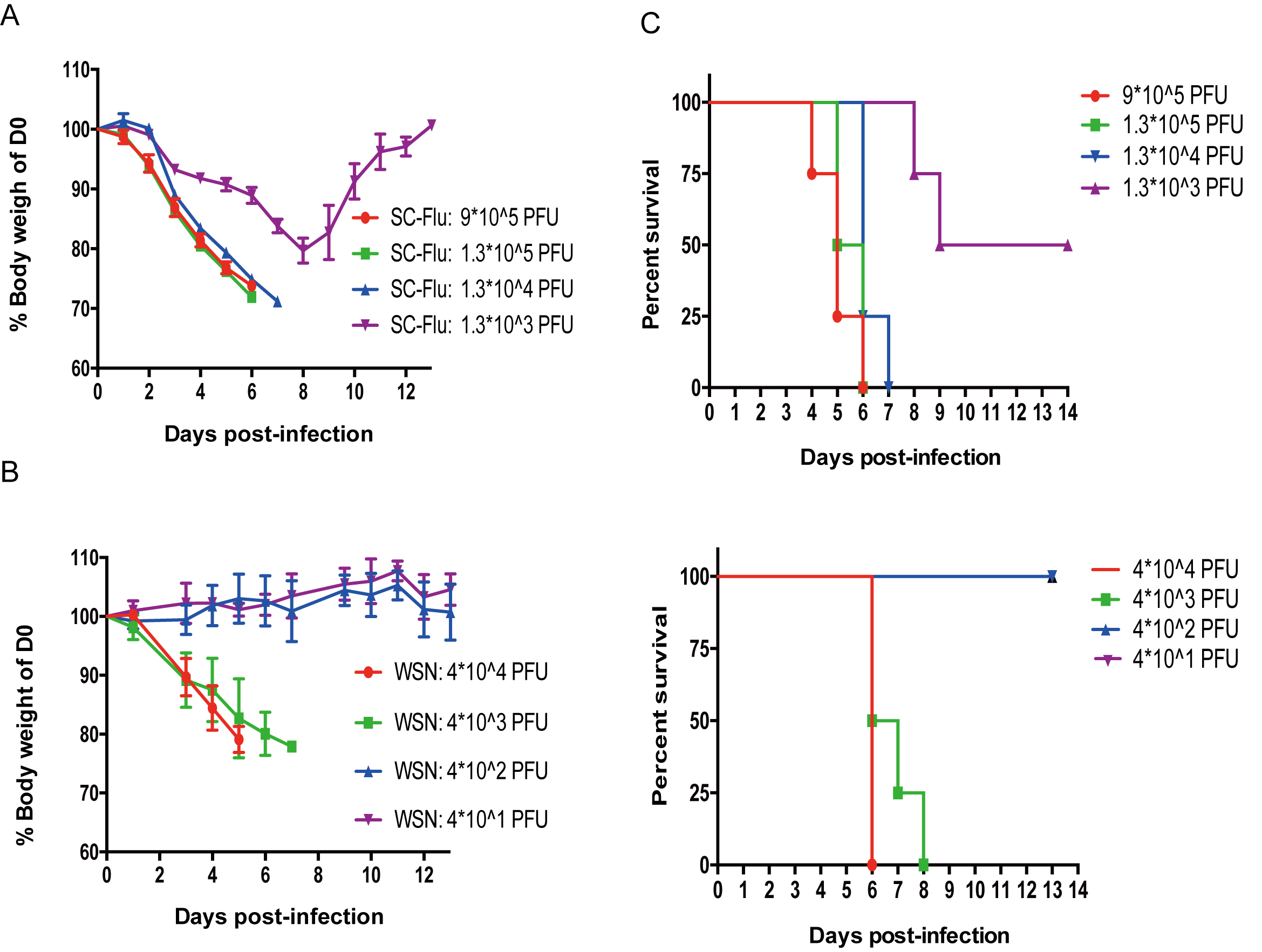
Figure 1. Exploration of median lethal dose of WSN and SC09. Mice were divided into four groups randomly and different doses of H1N1 were used to infect mice. The body weights of mice were monitored throughout the infection time course from Day 0 to Day 14 (A and C). The survival curve of mice is shown in B and D.
- Mice infection with different doses of virus for different purposes
To explore the impact of TCP on mice body-weight loss and mortality, we infected mice with a lethal dose of viruses to see if treatment of TCP can rescue or worsen the disease. For this, we infected the mice with a dose of 10,000 pfu of WSN (about 5LD50) virus to guarantee a lethal infection. (Figures 4A and 4B of Shan et al., 2017)- Transfer the mice (BALB/c mice aged 6-8 weeks) to ABSL2 for adaptive feeding for 3-7 days.
- Divide the mice (BALB/c mice aged 6-8 weeks) randomly into four groups: PBS-mock group, TCP-mock group, PBS-WSN treated group, and TCP-WSN treated group.
- Dilute 10,000 pfu of WSN in 50 μl PBS.
- Anesthetize mice with isoflurane and infect the mice intranasally by 50 μl droplets containing different concentrations of viruses diluted in PBS.
- Inject 1 mg/ml TCP (100 μl/mouse) and PBS (100 μl/mouse) intraperitoneally once a day for 10 days.
- Monitor the weight and death of the mice throughout the infection time course from Day 0 to Day 14 (Figures 4A and 4B of Shan et al., 2017).
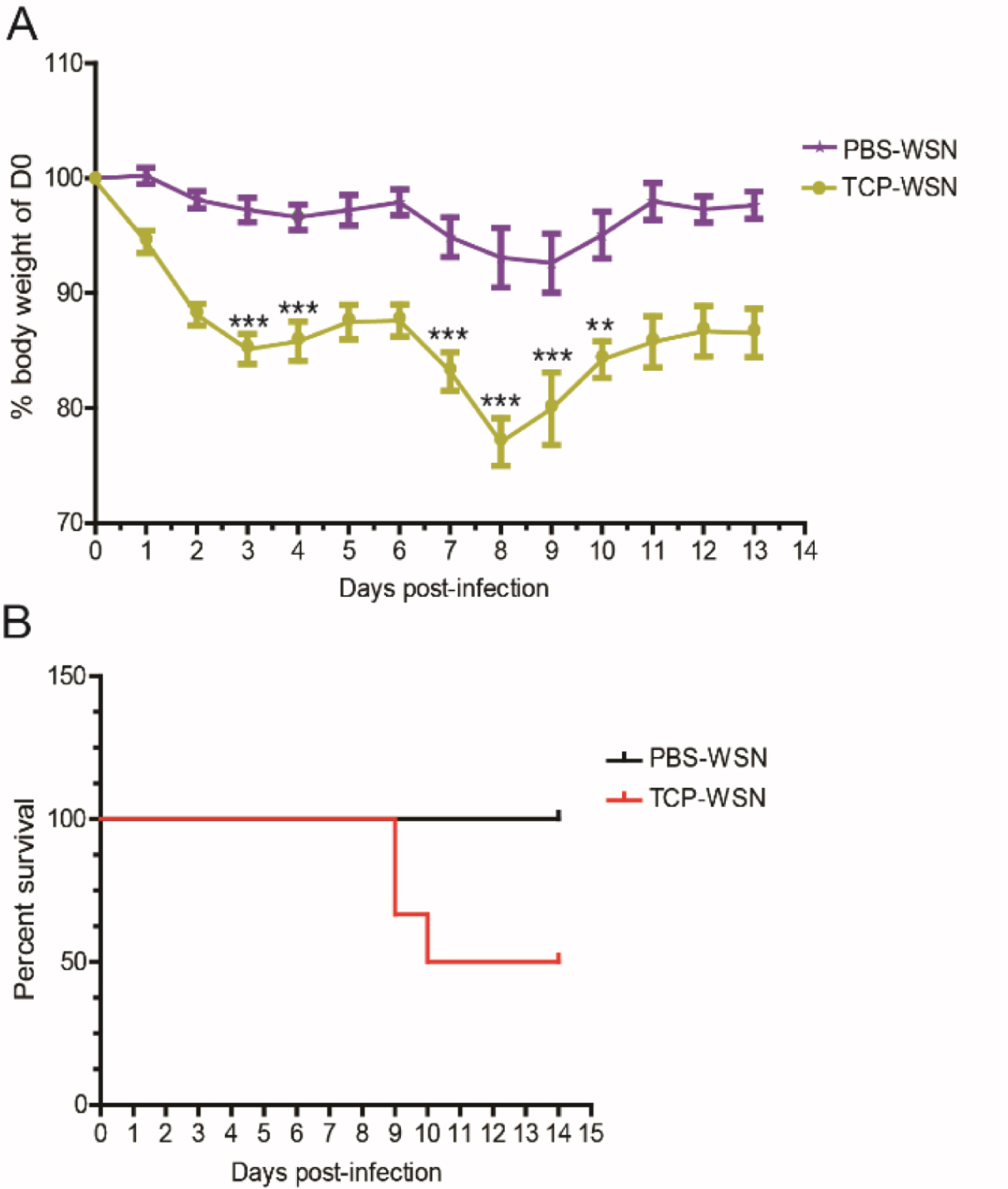
Figure 2. Low doses of viruses were used to guarantee mice of all groups could survive till at least Day 9 post-infection. The body weights of mice were monitored throughout the infection time course from Day 0 to Day 14 (A). The survival curve of mice is shown in B.
Data analysis
The median lethal dose is calculated with Bliss algorithm using Statistical Product and Service Solutions (SPSS). Survival curve and weight change curve are analyzed using GraphPad Prism. Data are analyzed by Student's t-test. P value of < 0.05 is considered to be statistically significant difference. ns, P >0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical analyses are done using GraphPad Prism.
Recipes
- 1 mg/ml Trans-2-phenylcyclopropylamine hydrochloride TCP
Dissolve 50 mg of TCP in 50 ml PBS
Once reconstituted, the solution should be stored at 4 °C, protected from light - 2.4% Avicel
Dissolve 1.2 g of Avicel in 50 ml 18 MΩ H2O
Once reconstituted, the solution is autoclaved and then stored at 4 °C - 6% BSA
Dissolve 0.6 g of BSA in 100 ml PBS
Once reconstituted, the solution is filtered with 0.22 μm filters and then stored at -20 °C - Overlay medium
1x DMEM
1.2% (w/v) Avicel
0.3% (w/v) BSA
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China for Distinguished Young Scholars (31525008) to B.L. and the National Natural Science Foundation of China (81590766, 81330072, 81360250, 31500714, 31370863, 81772202, 31570171); Chinese National Program on Key Basic Research Project Grants 2014CB541803 and 2014CB541903; The National Key Research and Development Program of China (2016YFC1200403); “Shanghai Academic Research Leader Program” (16XD1403800) to B.L. and “Rising Star Talent Program” (15QA1404000) to K.X. from the Science and Technology Commission of Shanghai Municipality.
Competing interests
The authors have declared that no competing interests exist.
Ethics
The animal experiments were approved by the Institutional Animal Care and Use Committee of the Institut Pasteur of Shanghai, Chinese Academy of Sciences (Animal protocol #A2015006). All animal care and use protocols were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China.
References
- Hoffmann, E., Neumann, G., Kawaoka, Y., Hobom, G. and Webster, R. G. (2000). A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97(11): 6108-6113.
- Kumar, A., Meldgaard, T. S. and Bertholet, S. (2018). Novel Platforms for the Development of a Universal Influenza Vaccine. Front Immunol 9: 600.
- Lim, B. H. and Mahmood, T. A. (2011). Influenza A H1N1 2009 (Swine Flu) and Pregnancy. J Obstet Gynaecol India 61(4): 386-393.
- Matrosovich, M., Matrosovich, T., Garten, W. and Klenk, H. D. (2006). New low-viscosity overlay medium for viral plaque assays. Virol J 3: 63.
- Molinari, N. A., Ortega-Sanchez, I. R., Messonnier, M. L., Thompson, W. W., Wortley, P. M., Weintraub, E. and Bridges, C. B. (2007). The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25(27): 5086-5096.
- Shan, J., Zhao, B., Shan, Z., Nie, J., Deng, R., Xiong, R., Tsun, A., Pan, W., Zhao, H., Chen, L., Jin, Y., Qian, Z., Lui, K., Liang, R., Li, D., Sun, B., Lavillette, D., Xu, K. and Li, B. (2017). Histone demethylase LSD1 restricts influenza A virus infection by erasing IFITM3-K88 monomethylation. PLoS Pathog 13(12): e1006773.
- Smith, G. J., Vijaykrishna, D., Bahl, J., Lycett, S. J., Worobey, M., Pybus, O. G., Ma, S. K., Cheung, C. L., Raghwani, J., Bhatt, S., Peiris, J. S., Guan, Y. and Rambaut, A. (2009). Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459(7250): 1122-1125.
- Thangavel, R. R. and Bouvier, N. M. (2014). Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410: 60-79.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhao, B., Shan, J., Xiong, R., Xu, K. and Li, B. (2018). H1N1 Virus Production and Infection. Bio-protocol 8(20): e3062. DOI: 10.21769/BioProtoc.3062.
Category
Microbiology > in vivo model > Viruses
Immunology > Animal model > Mouse
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









