- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Highly Sensitive Anion Exchange Chromatography Method for Measuring cGAS Activity in vitro
Published: Vol 8, Iss 20, Oct 20, 2018 DOI: 10.21769/BioProtoc.3055 Views: 6454
Reviewed by: Andrea PuharThomas Alexander PackardSaskia F. Erttmann

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of the Activity of Wildtype and Disease-Causing ALPK1 Mutants in Transfected Cells With a 96-Well Format NF-κB/AP-1 Reporter Assay
Tom Snelling
Nov 20, 2024 1602 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2277 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views
Abstract
Cyclic GMP-AMP synthase (cGAS) is a pattern recognition receptor (PRR) that senses double stranded DNA (dsDNA) in the cytosol and this leads to the activation of stimulator of interferon genes (STING) via the secondary messenger 2’3’-cyclic GMP-AMP (2’3’-cGAMP). STING then recruits TANK binding kinase 1 (TBK-1) and this complex can phosphorylate and activate interferon regulatory factor 3 (IRF3) leading to the induction of type I interferons and other antiviral genes. The cGAS:DNA complex catalyzes the synthesis of 2’3’-cGAMP and the purpose of the protocol presented here is to measure the in vitro activity of purified cGAS in the presence of dsDNA. The protocol was developed to elucidate the relationship between dsDNA length and the level of cGAS activity. The method involves an in vitro reaction with low concentrations of cGAS and dsDNA followed by quantification of the reaction product using anion exchange chromatography. The low concentrations of cGAS and dsDNA and the high sensitivity of this assay is a key advantage when comparing different DNA fragments’ ability to activate cGAS.
Keywords: Cyclic GMP-AMP synthaseBackground
The presence of double stranded DNA within the cytosol of a cell is a potential sign of infection by a DNA or retrovirus. The nucleotidyl transferase cGAS functions as a pattern recognition receptor that senses cytosolic dsDNA. cGAS is allosterically activated by dsDNA and catalyzes the conversion of ATP and GTP into the cyclic dinucleotide 2’3’-cGAMP (or simply cGAMP) (Ablasser et al., 2013; Civril et al., 2013; Diner et al., 2013; Gao et al., 2013; Kranzusch et al., 2013; Sun et al., 2013), which subsequently acts as a secondary messenger that induces an antiviral program in the infected cell. The active site of cGAS contains three acidic residues coordinating two magnesium ions. The role of these ions is to coordinate the triphosphate group of the donor nucleotide and the attacking hydroxyl group of the acceptor nucleotide. cGAS catalyzes the formation of cGAMP in two sequential steps. First, the triphosphate group of ATP is coordinated by the magnesium ions and the 2’-hydroxyl group of GTP makes a nucleophilic attack on the α-phosphate of ATP, which releases the β- and γ-phosphate as pyrophosphate. This leads to the formation of a noncanonical 2’,5’-phosphodiester linkage. The intermediate is then flipped around in the active site and now the triphosphate group of GTP is coordinated by the magnesium ions. This time the 3’-hydroxyl group of the AMP moiety makes the nucleophilic attack on the α-phosphate of GTP forming a 3’,5’-phosphodiester linkage (Civril et al., 2013; Gao et al., 2013; Hornung et al., 2014). Thus, the final product contains both a canonical and noncanonical phosphodiester linkage.
Not all dsDNA is equally efficient at activating cGAS. The minimum DNA length reported to activate cGAS in cells is 12 bp with guanosine overhangs (Herzner et al., 2015). However, the DNA’s ability to activate cGAS is strongly related to the length of the DNA. Increasing the DNA length leads to an increase in its ability to activate cGAS (Andreeva et al., 2017; Luecke et al., 2017). This effect is observed even when increasing the DNA length from 2 kb to 4 kb (Luecke et al., 2017). Furthermore, certain Y-form DNA generated during the reverse transcription of the HIV-1 genome is more potent at activating cGAS compared to conventional dsDNA of similar length (Herzner et al., 2015).
cGAMP acts as a secondary messenger that binds to the adaptor protein STING, and this leads to the induction of antiviral genes (Ablasser et al., 2013; Diner et al., 2013; Li et al., 2013; Sun et al., 2013; Zhang et al., 2013). STING is a transmembrane protein located in the endoplasmic reticulum (ER) membrane with a large C-terminal domain protruding into the cytosol (Ishikawa and Barber, 2008). When STING binds cGAMP, the complex moves to the Golgi apparatus and from there it moves to punctuated foci in the cytoplasm (Saitoh et al., 2009). The STING:cGAMP complex recruits TBK-1, and this leads to the phosphorylation of both STING and TBK-1. This phosphorylated complex can then phosphorylate and thereby activate IRF3, which then translocates to the nucleus where it induces the transcription of antiviral genes including type I interferons (Ishikawa et al., 2009; Tanaka and Chen, 2012). The STING:cGAMP complex will also activate nuclear factor kappa B (NFκB) transcription factors (Abe and Barber, 2014).
The method described in this protocol was used to show that the in vitro activation of recombinant human cGAS truncated to amino acids 155-522 (cGAS [155-522]) is dependent on DNA length. The tested interval of DNA lengths varied from 100 bp to 4,000 bp (Luecke et al., 2017). This method offers an alternative to thin layer chromatography (TLC)-based assays with radiolabeled ATP. Due to poor sensitivity, TLC-based assays normally use concentrations of both dsDNA and cGAS well above physiologically realistic concentrations. The advantage of using the protocol presented here is that no radioactivity or labeling of the substrates are needed and that the high sensitivity of this method makes it possible to use very low concentrations of both cGAS and dsDNA. In this protocol, the concentration of cGAS is ten-fold lower compared to classical TLC assays and we have avoided oversaturating the reaction with DNA. We use 1 ng/μl of dsDNA corresponding to 1.646 x 10-6 M bp. Assuming that one cGAS molecule covers approx. 20 bp (Andreeva et al., 2017), then 1.646 μM bp corresponds to 82.32 nM cGAS binding sites. Under this assumption, there is enough DNA to occupy about 82% of the cGAS used in this protocol. The use of low and approx. equimolar concentrations of cGAS and DNA (measured in cGAS binding sites) is important if you test DNA with small differences in affinity for cGAS. The impact of different affinities might be diminished if for example the DNA concentration is substantial above the saturation point.
This protocol allows for easy and robust quantifications of the cGAS product and compare reaction conditions (such as different buffers, DNA structures, DNA lengths, and cGAS preparations) but it is more time consuming than TLC when running multiple samples. The method described in this protocol was developed from a method designed to measure the activity of the oligoadenylate synthetase (OAS) proteins (Turpaev et al., 1997).
Materials and Reagents
- 1 ml single-use syringes (CHIRANA T. Injecta, catalog number: CH03001L )
- 100 ml and 500 ml GL45 thread reagent bottles including screw caps (SIMAX, catalog numbers: 1632414321100 and 1632414321500 )
- 50 ml tubes (SARSTEDT, catalog number: 62.547.254 )
- Autoclaved 1.5 ml tubes (BRAND, catalog number: 780500 )
- Cellulose acetate filter membranes 0.22 μm pore size (Frisenette, catalog number: CA047022 )
- Disposable nitrile gloves
- PCR tubes (VWR, catalog number: 211-0338 )
- Pipette tips with barrier (Thermo Fisher Scientific, ARTTM)
- Serological pipettes 10 ml (Th. Geyer, Labsolute, catalog number: 7695553 )
- dsDNA diluted to a concentration of 5 ng/μl in water or buffer NE (NucleoSpin® Gel and PCR Clean-up) (MACHEREY-NAGEL, catalog number: 740609 )
Note: If agarose gel purification of the DNA is desired, use NucleoSpin® Gel and PCR Clean-up for extraction of the DNA (MACHEREY-NAGEL, catalog number: 740609 ). - Ice
- 100 mM ATP (Thermo Fisher Scientific, catalog number: R0441 )
- 100 mM GTP (Thermo Fisher Scientific, catalog number: R0461 )
- Concentrated hydrochloric acid (Sigma-Aldrich, catalog number: 30721-1L )
- Magnesium chloride hexahydrate (Sigma-Aldrich, catalog number: M2670-1KG )
- Sodium hydroxide (VWR, catalog number: 28240.292 )
- Sodium chloride (VWR, catalog number: 27810.295 )
- Tris (VWR, catalog number: 103156X )
- Ultrapure water 18.2 MΩ obtained from PURELAB Chorus 1 (Elga Veolia)
- Zinc chloride (VWR, catalog number: 29156.231 )
- Glycerol (VWR, catalog number: 24388.295 )
- HEPES (VWR, catalog number: 30487.297 )
- 2 μM purified cGAS [155-522] stock (see Recipes)
- MgCl2 (200 mM) (see Recipes)
- ZnCl2 (10 mM) (see Recipes)
- NaOH (5 mM) (see Recipes)
- Tris (pH 7.5, 1 M) (see Recipes)
- 5x reaction buffer (see Recipes)
- Buffer A (see Recipes)
- Buffer B (see Recipes)
- ATP (10 mM) (see Recipes)
- GTP (10 mM) (see Recipes)
Equipment
- 2 ml sample loop for ÄKTApurifier 10 (GE Healthcare, catalog number: 18111402 )
- ÄKTApurifier 10 (GE Healthcare)
- Aluminum cooling block for PCR tubes (e.g., Sigma-Aldrich, catalog number: Z740270-1EA )
- -80 °C freezer
- Vacuum pump
- Centrifuge for 1.5 ml tubes (Eppendorf, model: MiniSpin® , catalog number: 5452000018)
- Injection needle for ÄKTApurifier 10 (GE Healthcare, catalog number: 18180142 )
- Laboratory balance with a readability of 0.001 g
- Microcentrifuge for PCR tubes (SpectrafugeTM Mini) (Sigma-Aldrich, Labnet International, catalog number: S7816EU-1EA )
- pH electrode (VWR, catalog number: 662-1157 )
- pH meter (VWR, catalog number: 662-1421 )
- Pipetboy
- Pipettes (Finnpipette, Thermo Fisher Scientific)
- RESOURCE Q 1 ml (GE Healthcare, catalog number: 17117701 )
- Thermal Cycler PCR machine (Bio-Rad Laboratories, model: T100TM )
- Vacuum filter funnel for GL45 threaded reagent bottles and 47 mm filter membrane diameter, e.g., NalgeneTM Polysulfone Reusable Bottle Top Filter (Thermo Fisher Scientific, catalog number: DS0320-5045 )
Software
- Unicorn 5.11 AA or 7 (GE Healthcare, catalog numbers: 28400955 or 29203853)
Procedure
Note: Gloves should be worn during all steps of this protocol to protect your samples from phosphatase contamination.
- Preparing control samples for assessing quality and elution profile of ATP, GTP and cGAMP
Note: Keep everything on ice for this step.- Mix the ATP sample, GTP sample, and cGAMP sample as described below in 1.5 ml tubes.
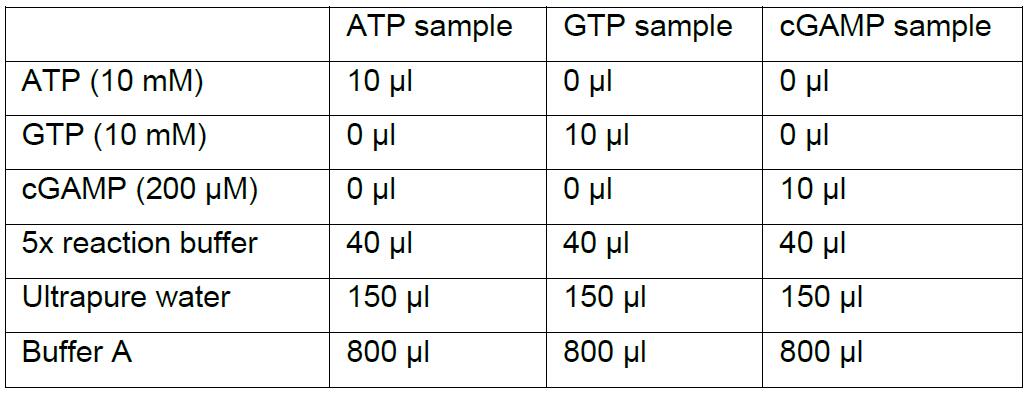
- The samples can be stored at -80 °C and thawed immediately before centrifugation and subsequent analysis on the RESOURCE Q 1 ml column (see Procedure D).
- Mix the ATP sample, GTP sample, and cGAMP sample as described below in 1.5 ml tubes.
- Dilution of DNA fragments and preparation of enzyme master mix
Notes:- Procedures B and C in this protocol are performed without pausing. Place the aluminum cooling block on ice for 30 min before starting Procedure B and keep it on ice throughout Procedures B and C.
- Keep everything on ice during this step, mix the enzyme master mix on ice, and keep the enzyme master mix on ice.
- Dilute the DNA fragments you are testing to a concentration of 5 ng/μl in ultrapure water or buffer NE.
- Mix enzyme master mix for the desired number of reactions according to the table below.
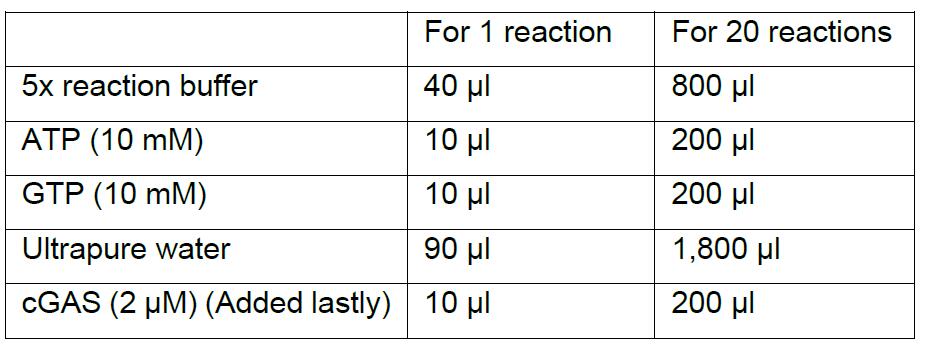
Notes:- Enzyme master mix can be prepared for any number of reactions simply by upscaling the recipe. It is highly recommended that you make a surplus of the enzyme master mix. For example, if you want to make four reactions you should multiply the recipe for one reaction by five.
- The number of reactions that you will make is the number of DNA species you want to test plus a negative control with no DNA.
- Mix the master mix well by pipetting carefully up and down several times using a pipette with a suitable volume. Do not introduce air bubbles or make it foam!
- In vitro cGAS reaction
- Immediately after preparing the enzyme master mix, place one PCR tube per reaction into the aluminum cooling block (remember a tube for the DNA-free negative control).
- Pipette 160 μl enzyme master mix into each PCR tube.
- Add 40 μl of a given DNA species/fragment (5 ng/μl) into a corresponding PCR tube and pipette up and down ten times to ensure mixing (avoid air bubbles).
- For the negative control, add 40 μl of ultrapure water or buffer NE depending on what the DNA fragments are suspended in.
Note: Addition of the DNA to the PCR tubes should be done swiftly but without forming foam in the samples. The cGAS catalyzed reaction is not occurring to any noticeable extent while the samples are kept on ice. - Centrifuge the PCR tubes for 30-60 s on the small SpectrafugeTM to remove any drops sitting on the side of the tube and to remove any air bubbles.
- Place the PCR tubes back in the cooling block.
- The T100TM Thermal Cycler PCR machine is programmed to 2 h at 37 °C, 10 min at 95 °C, and 12 °C for indefinite. The lid heating is set to 105 °C and the sample volume is set to maximum (100 μl).
- Transfer the PCR tubes to the PCR machine and start the program.
- When the 2 h at 37 °C and 10 min at 95 °C has passed, move the PCR tubes to the -80 °C freezer, where they are stored until the analysis (Procedure D).
- Analyzing samples on RESOURCE Q 1 ml
Note: The reactions are analyzed one at a time. The ATP, GTP, and cGAMP samples prepared in Procedure A are analyzed by continuing from Step D3.- Thaw a single PCR tube and transfer all 200 μl to a 1.5 ml tube.
- Add 800 μl buffer A to the 1.5 ml tube and mix well.
- Centrifuge the now diluted reaction sample at 12,100 x g for 15 min at 4 °C.
- Prepare the ÄKTApurifier 10 and RESOURCE Q 1 ml column.
- Wash pump A in buffer A and pump B in buffer B.
- Wash the entire flow path including the 2 ml sample loop in buffer A with a flow of 1 ml/min until the conductivity is stable and below 2.5 mS/cm.
- Connect the RESOURCE Q 1 ml column to the system and equilibrate the column in buffer A.
- Fix the injection needle to a 1 ml syringe and draw 900 μl of the diluted reaction sample into the syringe without disturbing the pellet that might have appeared after centrifugation.
- Inject 800 μl onto the sample loop.
- Use the Unicorn software to program the ÄKTApurifier 10 to do the following (see Table S1 for full variable list):
Program:
Wave length 1 = 280 nm
Wave length 2 = 254 nm
Wave length 3 = 215 nm (optional)
Flow rate: 0.5 ml/min
Equilibrate with 2 column volumes buffer A
Empty loop with 10 ml
Linear gradient from 0% to 50% buffer B over 25 column volumes
Wash column with 100% buffer B over 5 column volumes
Re-equilibrate with 5 column volumes buffer A
No fractionation
Note: The maximal pressure for the column is 1.5 MPa but the pressure generated by the column during this application is usually less than 0.5 MPa. We run the chromatography at 4 °C but it will also work at room temperature. - When the run is finished the next reaction sample can be analyzed by repeating Procedure D.
Data analysis
Open the Unicorn 5 evaluation window (If you are using Unicorn 7, use the Evaluation Classic application) and open the data you wish to analyze (data from each anion exchange chromatography run can be found in the Result Navigator in the left side of the evaluation window). When the data is open click “Integrate” and choose “Peak Integrate” from the drop down menu.
A new window opens. In this window, there will be two lists on white background. In the left list choose the 254 nm UV for integration (if the program is as described in supplementary Figure 1 the 254 nm UV is the second element from the top and when the list element is highlighted in blue it is chosen). The baseline should be set to “Calculate Baseline” (default). Click “OK” and a peak table appears below the curves. The identified peaks are listed according to retention volume and you can read the area under the curve (AUC) for the cGAMP peak and for any other peak in the chromatogram. Make sure that the calculated baseline looks correct. If the curve has abrupt and discontinuous changes around the peaks due to air in the system or other artifacts, it can give an unreliable baseline and unreliable results. If the curve is discontinuous, it might be necessary to repeat the experiment.
The AUC has the unit mAU•ml and for the cGAMP peak the AUC is a measurement for the amount of cGAMP eluting from ion exchange column. The amount of cGAMP eluting from the column is dependent on the amount of cGAMP produced in a reaction. For this reason, it is possible to use the AUC of the cGAMP peak to represent the activity of cGAS. The AUC can for example be presented in a column bar graph or a column scatter plot.
Note: It is possible to convert the quantification of cGAMP from mAU•ml to nmol. This requires that you make a cGAMP standard curve by running different concentrations of cGAMP on the anion exchange column.
ATP gives a peak at a conductivity of approx. 17.1 mS/cm. There might also be a small ADP peak at a conductivity of approx. 13.6 mS/cm. GTP gives a peak at a conductivity of approx. 18.4 mS/cm. There might also be a small GDP peak at a conductivity of approx. 14.9 mS/cm. 2’3’-cGAMP gives a peak at a conductivity of approx. 9.9 mS/cm.
There can be small variations between runs and between ÄKTApurifier systems. For representative data, see Figure 1 and Luecke et al. (2017).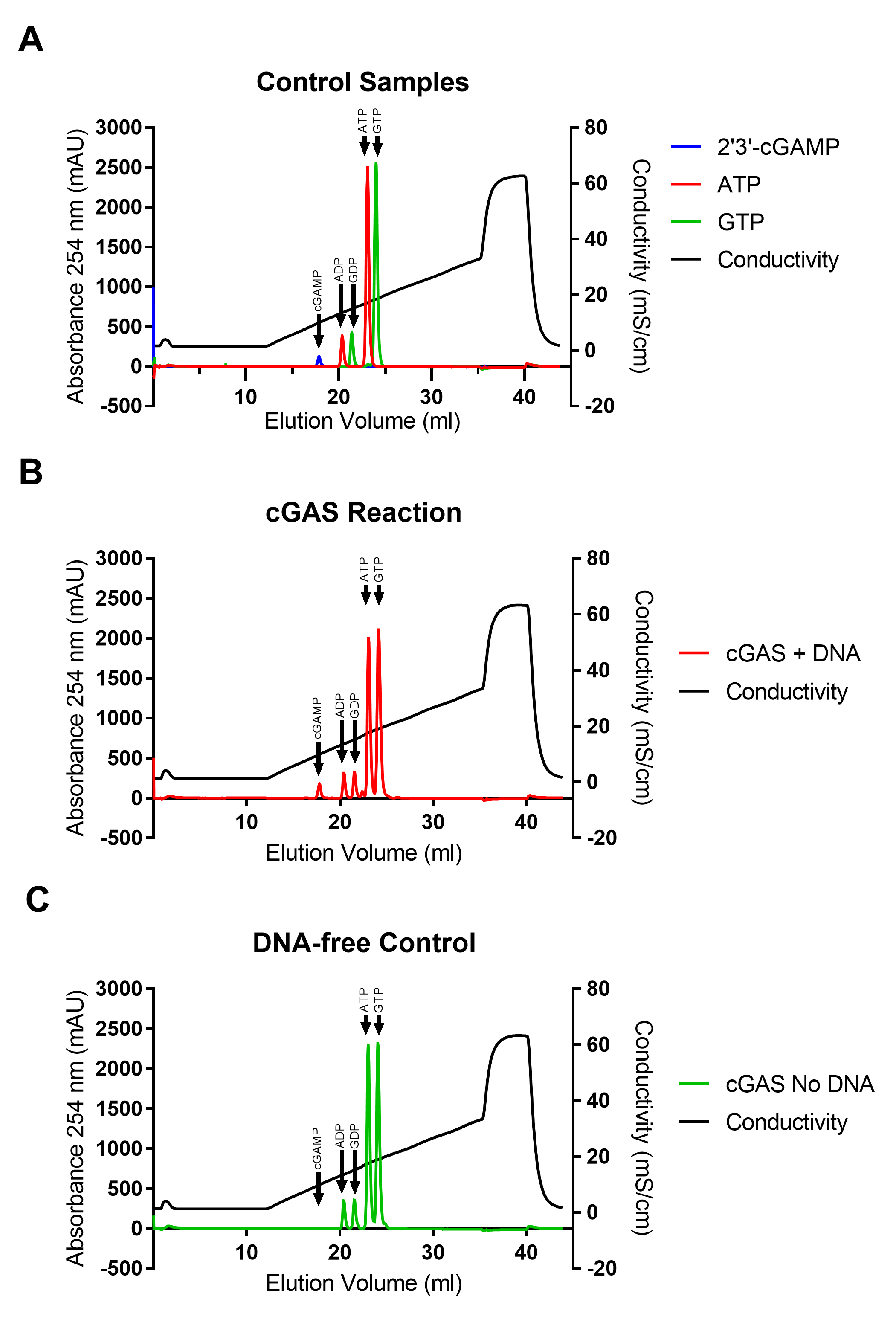
Figure 1. Examples of chromatograms. A. Chromatograms of 2’3’-cGAMP, ATP, and GTP. B and C. Chromatograms of two different reactions. B) cGAS with a 4 kb PCR fragment. C) cGAS without DNA. The data has previously been published in Luecke et al. (2017).
Notes
The NTP’s are very sensitive to dephosphorylation. It is therefore very important to protect the samples from phosphatases from the environment. That is why gloves should be worn when working with or handling the samples and reagents. The 1.5 ml tubes should be autoclaved and in general care should be taken not to contaminate the samples with phosphatases.
Other reaction conditions suitable for cGAS can also be used in this assay. Avoid chelating agents such as EDTA in the buffers as they interfere with the anion exchange column. In our experience, high salt concentrations can also inhibit the reaction. If you increase the amount of cGAS and DNA, be careful that you do not experience substrate depletion. We recommend that a minimum of 60% of the substrate is remaining after terminating the reaction
Recipes
- 2 μM purified cGAS [155-522] stock
2 μM purified human cGAS truncated to amino acids 155-522 suspended in 70 mM NaCl, 10% (v/v) glycerol, 20 mM HEPES, pH 7.5
Note: For cGAS purification see Luecke et al. (2017). E. coli was transformed with a pET-21a plasmid encoding cGAS [155-522] with an N-terminal Hexa His-MBP tag and a Tobacco Etch Virus (TEV) protease cleavage site. The fusion protein was expressed by the bacterial cells and the MBP tag was cleaved off during purification leaving four residues Gly-Ala-Met-Gly in front of Arg155. Dialysis can be used for changing the buffer of cGAS to 70 mM NaCl, 10% (v/v) glycerol, 20 mM HEPES, pH 7.5 before adjusting the concentration to 2 μM. - MgCl2 (200 mM)
Dissolve 2.03 g MgCl2•6H2O in 50 ml ultrapure water - ZnCl2 (10 mM)
Dissolve 0.682 g ZnCl2 in 500 ml ultrapure water - NaOH (5 M)
Dissolve 100 g NaOH in 500 ml ultrapure water - Tris (pH 7.5 1 M)
- Dissolve 60.57 g Tris in 400 ml ultrapure water and adjust the pH to 7.5 at room temperature using concentrated HCl
- Adjust the volume to 500 ml using ultrapure water
- 5x reaction buffer
- Mix 1.46 g NaCl with 12.5 ml 200 mM MgCl2, 0.25 ml 10 mM ZnCl2, 10 ml 1 M Tris pH 7.5
- Adjust the volume to 50 ml using ultrapure water
- Filtrate the 5x reaction buffer with a 0.22 μm cellulose acetate filter
- Store at -20 °C
- Buffer A
- Mix 10 ml 1 M Tris pH 7.5 with 400 ml ultrapure water
- Adjust the pH to 7.5 at room temperature using 5 M NaOH
- Adjust the volume to 500 ml using ultrapure water
- Filtrate the buffer with a 0.22 μm cellulose acetate filter
- Prepare the buffer fresh
- Buffer B
- Mix 22 g NaCl and 10 ml 1 M Tris pH 7.5 with 400 ml ultrapure water
- Adjust the pH to 7.5 at room temperature using 5 M NaOH
- Adjust the volume to 500 ml using ultrapure water
- Filtrate the buffer with a 0.22 μm cellulose acetate filter
- Prepare the buffer fresh
- ATP (10 mM)
Mix 100 μl 100 mM ATP with 900 μl ultrapure water
Make aliquots of a size that suits your requirement and store at -80 °C
Note: Be careful not to contaminate with phosphatases from your hands or the surroundings. - GTP (10 mM)
Mix 100 μl 100 mM GTP with 900 μl ultrapure water
Make aliquots of a size that suits your requirement and store at -80 °C
Note: Be careful not to contaminate with phosphatases from your hands or the surroundings.
Acknowledgments
We thank Professor Søren R. Paludan and Stefanie Luecke, Ph.D. for helping with the development of the method described in this protocol. We thank Hans Henrik Gad, Ph.D. for comments on the manuscript.
This work was funded by The Novo Nordisk Foundation (NNF17OC0028184) and the Danish Council for Independent Research, Natural Science (4181-00012B).
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Abe, T. and Barber, G. N. (2014). Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol 88(10): 5328-5341.
- Ablasser, A., Goldeck, M., Cavlar, T., Deimling, T., Witte, G., Rohl, I., Hopfner, K. P., Ludwig, J. and Hornung, V. (2013). cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature 498(7454): 380-384.
- Andreeva, L., Hiller, B., Kostrewa, D., Lassig, C., de Oliveira Mann, C. C., Jan Drexler, D., Maiser, A., Gaidt, M., Leonhardt, H., Hornung, V. and Hopfner, K. P. (2017). cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549(7672): 394-398.
- Civril, F., Deimling, T., de Oliveira Mann, C. C., Ablasser, A., Moldt, M., Witte, G., Hornung, V. and Hopfner, K. P. (2013). Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498(7454): 332-337.
- Diner, E. J., Burdette, D. L., Wilson, S. C., Monroe, K. M., Kellenberger, C. A., Hyodo, M., Hayakawa, Y., Hammond, M. C. and Vance, R. E. (2013). The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 3(5): 1355-1361.
- Gao, P., Ascano, M., Wu, Y., Barchet, W., Gaffney, B. L., Zillinger, T., Serganov, A. A., Liu, Y., Jones, R. A., Hartmann, G., Tuschl, T. and Patel, D. J. (2013). Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153(5): 1094-1107.
- Herzner, A. M., Hagmann, C. A., Goldeck, M., Wolter, S., Kübler, K., Wittmann, S., Gramberg, T., Andreeva, L., Hopfner, K. P., Mertens, C., Zillinger, T., Jin, T., Xiao, T. S., Bartok, E., Coch, C., Ackermann, D., Hornung, V., Ludwig, J., Barchet, W., Hartmann, G. and Schlee, M. (2015). Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16(10): 1025-1033.
- Hornung, V., Hartmann, R., Ablasser, A. and Hopfner, K. P. (2014). OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol 14(8): 521-528.
- Ishikawa, H. and Barber, G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455(7213): 674-678.
- Ishikawa, H., Ma, Z. and Barber, G. N. (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461(7265): 788-792.
- Kranzusch, P. J., Lee, A. S., Berger, J. M. and Doudna, J. A. (2013). Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep 3(5): 1362-1368.
- Li, X., Shu, C., Yi, G., Chaton, C. T., Shelton, C. L., Diao, J., Zuo, X., Kao, C. C., Herr, A. B. and Li, P. (2013). Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39(6): 1019-1031.
- Luecke, S., Holleufer, A., Christensen, M. H., Jonsson, K. L., Boni, G. A., Sorensen, L. K., Johannsen, M., Jakobsen, M. R., Hartmann, R. and Paludan, S. R. (2017). cGAS is activated by DNA in a length-dependent manner. EMBO Rep 18(10): 1707-1715.
- Saitoh, T., Fujita, N., Hayashi, T., Takahara, K., Satoh, T., Lee, H., Matsunaga, K., Kageyama, S., Omori, H., Noda, T., Yamamoto, N., Kawai, T., Ishii, K., Takeuchi, O., Yoshimori, T. and Akira, S. (2009). Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106(49): 20842-20846.
- Sun, L., Wu, J., Du, F., Chen, X. and Chen, Z. J. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339(6121): 786-791.
- Tanaka, Y. and Chen, Z. J. (2012). STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5(214): ra20.
- Turpaev, K., Hartmann, R., Kisselev, L. and Justesen, J. (1997). Ap3A and Ap4A are primers for oligoadenylate synthesis catalyzed by interferon-inducible 2-5A synthetase 1. FEBS Lett 408(2): 177-181.
- Zhang, X., Shi, H., Wu, J., Zhang, X., Sun, L., Chen, C. and Chen, Z. J. (2013). Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51(2): 226-235.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Holleufer, A. and Hartmann, R. (2018). A Highly Sensitive Anion Exchange Chromatography Method for Measuring cGAS Activity in vitro. Bio-protocol 8(20): e3055. DOI: 10.21769/BioProtoc.3055.
Category
Immunology > Host defense > General
Biochemistry > Other compound > cGAMP
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









