- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation and Purification of Proteins Secreted from Phytophthora sojae
Published: Vol 8, Iss 20, Oct 20, 2018 DOI: 10.21769/BioProtoc.3045 Views: 7013
Reviewed by: Zhibing LaiJuan DuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Hyphal Branching Assay Using Rhizophagus irregularis
Takaya Tominaga and Hironori Kaminaka
Aug 20, 2024 2362 Views

Silencing Arbuscular Mycorrhizal Fungal Gene Using Chitosan Nanoparticle-Mediated dsRNA Delivery System
Chumei Yan [...] Xianan Xie
Jun 5, 2025 2669 Views

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1601 Views
Abstract
Phytophthora sojae, the causal agent of soybean root and stem rot, is responsible for enormous economic losses in soybean production. P. sojae secrets various effectors to reprogram host immunity. The plant apoplastic space is a major battleground in plant-pathogen interactions. Here we describe a protocol for purification and isolation of secreted proteins from P. sojae, including precipitation of secreted proteins from P. sojae culture filtrate, chromatographic purification of the secreted proteins and analysis of the proteins by Mass spectrometry. With this protocol, it will be easier to identify potential apoplastic effectors in Phytophthora and will benefit our understanding of plant-microbe interactions.
Keywords: Phytophthora sojaeBackground
Purification of proteins secreted from Phytophthora species is essential for understanding Phytophthora pathogenesis. In the past, V8 juice and Plant (tomato, and Lima bean) juice medium have been used to culture Phytophthora and the culture filtrated was used for the subsequent analysis of Phytophthora secreted proteins. The drawback of these protocols is the culturing media contain huge amount of plant proteins, which represent a large proportion of the detected proteins. In this protocol, we made use of the Synthesis Liquid Medium, which does not contain any proteins. This medium significantly reduces the background of the Phytophthora culturing filtrate. Furthermore, making use of the Gel Filtration desalting and sieving columns instead of Ion Exchange columns, allows efficient and large-scale purification of Phytophthora secreted proteins.
Materials and Reagents
- Pipette tips
- Scalpel
- Filter paper (Whatman filters) (No. 1)
- 0.22 μm Millex-GV filter (Merck, catalog number: SLGP033NS )
- Amicon Ultra-15 Centrifugal Filter Unit 3KD (Merck, catalog number: UFC900308 )
- P. sojae strain (P6497)
- V8 juice (https://www.campbells.com/v8/vegetable-juice/)
- Calcium carbonate (CaCO3) (Sigma-Aldrich, catalog number: 398101 )
- Potassium Dihydrogen Phosphate (KH2PO4) (Merck, catalog number: 1048730250 )
- Yeast extract (Sigma-Aldrich, catalog number: Y1625-1KG )
- Magnesium sulfate heptahydrate (MgSO4•7H2O) (Sigma-Aldrich, catalog number: RES0089M-A702X )
- D-glucose (Sigma-Aldrich, catalog number: G8270-100G )
- L-asparagine (Sigma-Aldrich, catalog number: A0884-100G )
- β-sitosterol (Sigma-Aldrich, catalog number: S1270-10MG )
- Thiamine(VB1) (Sigma-Aldrich, catalog number: T4625-5G )
- Milli-Q-filtered H2O
- Ammonium sulfate (NH4)2SO4 (Sigma-Aldrich, catalog number: A4418-100G )
- Tris-aminomethane (Tris-HCl) (Sigma-Aldrich, catalog number: 154563-1KG )
- Ethylene Diamine Tetraacetic Acid (EDTA) (Sigma-Aldrich, catalog number: 80849-10MG )
- 30% acrylamide
- 10% sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771-100G )
- 10% ammonium persulfate (Sigma-Aldrich, catalog number: A3678-25G )
- N,N,N',N'-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281-25ML )
- Coomassie brilliant blue (CBB-R-250) (Beyotime Biotechnology, catalog number: P0017 )
- Ethanol (ALADDIN-E, catalog number: A112719 )
- Acetic acid (ALADDIN-E, catalog number: A116174 )
- Methanol (ALADDIN-E, catalog number: M116127 )
- V8 juice with 2% agar (see Recipes)
- Synthesis liquid medium (see Recipes)
- TE buffer (see Recipes)
- SDS-PAGE Gel (see Recipes)
- R-250 staining solution (see Recipes)
- Destaining solution (see Recipes)
Equipment
- Pipettes
- Conical flask (1 L)
- Beaker (5 L)
- Measuring cylinder (1 L)
- Constant temperature incubator (CTI) (25 °C)
- Constant temperature shaker (CTS) (25 °C, 100 rpm)
- Centrifuge
- Liquid chromatograph (GE Healthcare, model: ÄKTA avant 25 )
- Column Sephadex G-25S (GE Healthcare, catalog number: 17140801 )
- Chromatography column (GE Healthcare, model: SuperdexTM 200 Increase, catalog number: 28990944 )
Note: The materials, reagents and equipment not provided company and catalog number can be ordered from any qualified company for using in this experiment.
Procedure
- Preparation and culture of P. sojae for secreted protein
- P. sojae strain P6497 was grown in sterilized 10% V8 juice with 2% agar medium at 25 °C in the Constant Temperature incubator (CTI) under dark conditions for 4 days (Figure 1).

Figure 1. P. sojae colony grown on V8 plates for 4 days - For secreted proteins production, cut P. sojae grown on V8 plates into 3 mm x 3 mm discs with a sterilized scalpel. Transfer twenty P. sojae discs into 1 L synthesis liquid medium and incubate on the Constant temperature shaker (CTS) (25 °C, 100 rpm) for 10 days (Figure 2).

Figure 2. P. sojae cultured in synthesis liquid medium for 10 days
- P. sojae strain P6497 was grown in sterilized 10% V8 juice with 2% agar medium at 25 °C in the Constant Temperature incubator (CTI) under dark conditions for 4 days (Figure 1).
- Extraction and collection of secreted protein from medium by P. sojae
- Collect P. sojae culture fluid by centrifugation at 1,200 x g for 20 min and filter through two layers of Whatman filters (No. 1) and then a 0.22 μm Millex-GV filter.
- Add 70 g of (NH4)2SO4 per 100 ml filtrate for precipitation and incubate for 12 h at 4 °C.
- Dissolve the precipitate in 4 ml TE buffer.
- Chromatographic purification and isolation of extracts secreted by P. sojae (see the Video 1)Video 1. Gel filtration desalting chromatography for separating the precipitated proteins by AKTA
- Gel filtration desalting chromatography for separating the precipitated proteins
- Equilibrate the Sephadex G-25S, the bound material and elute system using TE buffer.
- Load the protein samples into the Column Sephadex G-25S.
- Collect the fractions corresponding to each absorbance peak respectively and concentrate using 3-kD molecular mass centrifugal filter devices. (The absorption peaks stand for the line of UV280 on the screen of AKTA, Figure 3)
- Gel filtration sieving chromatography
- Collect the eluted samples from Sephadex G-25S and subsequently inject into TE buffer-equilibrated SuperdexTM 200 Increase chromatography column.
- Collect the fractions corresponding to each absorbance peak respectively, and then desalt and concentrate using 3-kD molecular mass centrifugal filter devices (see Amicon Ultra-15 Centrifugal Filter Unit 3-kD manual).
- SDS-PAGE analysis of the fractions corresponding to each absorbance peak
- Load the concentrated protein (20 μl about 100 μg/ml) into 10% SDS-PAGE gel and subject to electrophoresis for 1 h in 100 V.
- Stain the gel with Coomassie Brilliant Blue overnight and then destain for 3 h at room temperature.
- Separation and elution of protein in SDS-PAGE Gel
- For the fractions showing more than one band after Coomassie Brilliant Blue staining in the SDS-PAGE gel (Figure 3), cut each protein band from gel and into 2-mm segments. Dry it (remove the water with Filter paper).
- After drying, dissolve these segments in 0.1 ml 0.1% SDS (pH 6) and incubate at 70 °C for 1 h and then 37 °C overnight.
- Collect the supernatant by centrifugation at 12,000 x g for 10 min at 4 °C.
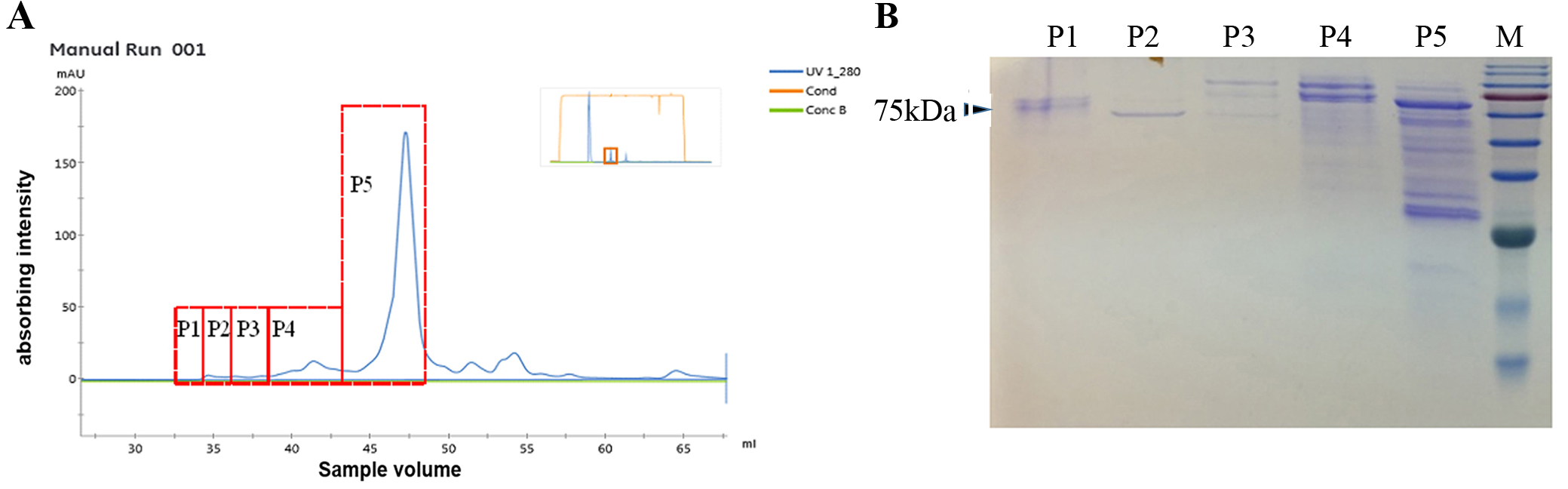
Figure 3. SDS-PAGE analysis of the protein fractions. A. Absorbance peaks corresponding to the fractions in B, respectively. B. Fractions of secreted protein show on SDS-PAGE gel after Coomassie Brilliant Blue staining.
- Gel filtration desalting chromatography for separating the precipitated proteins
- Identification of secreted protein by mass spectrometry
Analyze the collected supernatant by LC-MS/MS as described before (Wang et al., 2003; Ma et al., 2015). Proteins matched with peptides identified by mass spectrometry are used for further research.
Note: For further analysis of these proteins, they were expressed in Pichia pastoris (Km71) using the pPIZ9k vector or Escherichia coli (Rosetta) using pET32a.
Data analysis
Based on the molecular mass, secreted proteins were separated by Gel filtration sieving chromatography. Protein with similar molecular mass will form one peak. The area of the peak represents the abundance of purified proteins. Proteins collected in each peak were subsequently used for LC-MS/MS analysis and the data were shown in Ma et al. (2015).
Notes
- Kanamycin (50 μg/μl) can be added in the medium in case of bacterial pollution.
- (NH4)2SO4 should be added into the medium slowly with continuous swirling.
Recipes
- V8 juice with 2% agar (1 L)
V8 juice: 100 ml
CaCO3: 1 g
ddH2O: 900 ml
Agar power: 20 g - Synthesis liquid medium (1 L)
0.5 g KH2PO4
0.5 g Yeast extract
0.25 g MgSO4•7H2O
0.001 g thiamine
25 g Glucose
1 g Asparagines
0.01 g β-Sitosterol
Add water to a volume of 900 ml
Mix and make up to 1 L with Milli-Q-filtered H2O - TE buffer
10 mM Tris-HCl (pH 7.5) with 1 mM EDTA (TE) - 10% SDS-PAGE Gel
- Resolving gel buffer (10 ml):
4 ml Milli-Q-filtered H2O
3.3 ml 30% acrylamide (Bio-Rad)
2.5 ml 1.5 M Tris-HCl (Sigma-Aldrich) (pH 8.8)
0.1 ml 10% SDS (Sigma-Aldrich)
0.1 ml 10% ammonium persulfate (Sigma-Aldrich)
0.004 ml TEMED (Bio-Rad) - Stacking gel buffer (3 ml):
2.1 ml Milli-Q-filtered H2O
0.5 ml 30% acrylamide (Bio-Rad)
0.38 ml 1.5 M Tris-HCl (Sigma-Aldrich) (pH 6.8)
0.03 ml 10% SDS (Sigma-Aldrich)
0.03 ml 10% ammonium persulfate (Sigma-Aldrich)
0.003 ml TEMED (Bio-Rad)
- Resolving gel buffer (10 ml):
- R-250 staining solution (100 ml)
Methanol: 45 ml
ddH2O: 45 ml
Acetic acid: 10 ml
R-250 Coomassie: 0.25 g - Destaining solution (1 L)
Ethanol: acetic acid: ddH2O = 1:1:2
Acknowledgments
The authors would like to thank Dr. Zhenchuan Ma and this work was fund by grants to Yuanchao Wang from the China National Funds for Innovative Research Groups (Grant No. 31721004), the Special Fund for Agro-scientific Research in the Public Interest (201303018), and the China Agriculture Research System (CARS-004-PS14).
Competing interests
The authors declare that there is no conflict of interest.
References
- Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., Dong, S., Zhang, Z., Dou, D., Zheng, X., Tyler, B. M. and Wang, Y. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27(7): 2057-2072.
- Wang, Y. C., Hu, D. W., Zhang, Z. G., Ma, Z. C., Zheng, X. B. and Li, D. B. (2003). Purification and immunocytolocalization of a novel Phytophthora boehmeriae protein inducing the hypersensitive response and systemic acquired resistance in tobacco and Chinese cabbage. Physiol Mol Plant Pathol 63: 223-232.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xia, Y., Wang, Y. and Wang, Y. (2018). Preparation and Purification of Proteins Secreted from Phytophthora sojae. Bio-protocol 8(20): e3045. DOI: 10.21769/BioProtoc.3045.
Category
Microbiology > Microbe-host interactions > Fungus
Molecular Biology > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












