- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Real-time PCR Analysis of PAMP-induced Marker Gene Expression in Nicotiana benthamiana
Published: Vol 8, Iss 19, Oct 5, 2018 DOI: 10.21769/BioProtoc.3031 Views: 9304
Reviewed by: Wende Liu Satyabrata NandaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

3’ Rapid Amplification of cDNA Ends (3’ RACE) Using Arabidopsis Samples
Encarnación Rodríguez-Cazorla [...] Antonio Vera
Oct 5, 2015 21342 Views

RNA Stability Measurements Using RT-qPCR in Arabidopsis Seedlings
Tianran Jia and Brandon H. Le
Jul 20, 2020 6996 Views

Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
Valentín Luna-García and Stefan de Folter
Sep 5, 2024 2066 Views
Abstract
Perception of pathogen-associated molecular patterns (PAMPs) often triggers various innate immune responses in plants. The transcriptional changes of defense-related genes are often used as a marker to assay PAMP-triggered plant immune response. Here we described a protocol to monitor the relative expression level of marker genes in Nicotiana benthamiana upon treatment with PAMPs. The procedure includes leaf treatment using PAMPs, total RNA isolation, cDNA synthesis, quantitative real-time PCR and data analysis. This protocol is applicable to monitor marker gene expression triggered by different PAMPs in N. benthamiana.
Keywords: PAMPsBackground
Pathogen-associated molecular patterns, namely PAMPs, are a class of molecules derived from pathogens and are relatively conserved across microorganisms. Multiple PAMPs such as flg22 and XEG1 (Felix et al., 1999; Ma et al., 2015), have been characterized that can be detected by plant cell surface localized pattern-recognition receptors (PRRs) and thereby induce PAMP-triggered immunity (Couto and Zipfel, 2016). One of the predominant PAMP-triggered responses is the activation of defense-related maker genes (Navarro et al., 2004; Zipfel et al., 2006). Nicotiana benthamiana has been used extensively as a model plants and is sensitive to multiple PAMPs. In N. benthamiana, the marker genes, such as NbCYP71D20, NbACRE31 and NbWRKY22, were previously found that are rapidly activated upon PAMP treatment (Heese et al., 2007; Segonzac et al., 2011; Wang et al., 2018). Here, we describe a detailed protocol for checking the PAMP-triggered marker gene expression in N. benthamiana. The relative gene expression was also determined in parallel using a negative control to exclude the background noise.
Materials and Reagents
- MicroAmpTM Splash-Free 96-Well Base (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4312063 )
- 1,000 µl pipette tips (Corning, Axygen®, catalog number: TF-1000-R-S )
- 200 µl pipette tips (Corning, Axygen®, catalog number: TF-200-R-S )
- 10 µl pipette tips (Corning, Axygen®, catalog number: TF-300-R-S )
- 1 ml needless syringe (BD, catalog number: 309659 )
- 1.5 ml RNase-free tube (Corning, Axygen®, catalog number: MCT-150-C )
- Axygen® 0.2 ml Polypropylene PCR Tube Strips (8-Tubes/Strip) (Corning, Axygen®, catalog number: PCR-0208-C )
- MicroAmpTM Optical 96-Well Reaction Plate with Barcode (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4306737 )
- MicroAmpTM Optical Adhesive Film (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4360954 )
- Tissue paper
- Healthy 5-6 weeks N. benthamiana plants (see Figure 1)
- Any PAMPs of interest and corresponding control solution
- Liquid nitrogen
- β-mercaptoethanol (Solarbio, catalog number: M8210 )
- E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek, catalog number: R6834-01 )
- DNase/RNase-free ddH2O (Solarbio, catalog number: R1600 )
- PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Clontech, catalog number: RR047A )
- TB GreenTM Premix Ex TaqTM (Tli RNase H Plus) (Takara Bio, Clontech, catalog number: RR420A )
- 1 mM flg22 stock solution (Gene Script, RP19986) (see Recipes)
- 1 mM XEG1 stock solution (see Recipes)
- 70% ethanol (see Recipes)
Equipment
- Mortar and pestle
- Growth chamber
- NanoDropTM 1000 spectrophotometer (Thermo Fisher Scientific, model: NanoDropTM 1000 )
- Vortexer (Scientific Industries, model: Vortex-Genie 2 , catalog number: SI-0246)
- Cold Centrifuge (Eppendorf, model: 5424 R , catalog number: 5404000014)
- Pipettes 100-1,000 µl (Eppendorf, catalog number: 3120000062 )
- Pipettes 10-100 µl (Eppendorf, catalog number: 3120000046 )
- PCR Thermal Cyclers (Thermo Fisher Scientific, Applied BiosystemsTM, model: 2720, catalog number: ED000651 )
- MicroAmpTM Adhesive Film Applicator (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4333183 )
- MPS 1000 Mini PCR Plate Spinner (Labnet International, catalog number: C1000 )
- ABI 7500 fast real-time PCR system (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4351107 )
Software
- Applied Biosystems Sequence Detection Software v1.4.0
Procedure
- Infiltration of Nicotiana benthamiana
- N. benthamiana plants were grown in a growth chamber at 19-22 °C under long-day conditions (14-h-light/10-h-dark) and 70-80% relative humidity for 5-6 weeks as shown in Figure 1.

Figure 1. Nicotiana benthamiana plant used for PAMP treatment - N. benthamiana leaves of the same position and similar size were treated with PAMPs (e.g., flg22) or control (e.g., ddH2O) by infiltration using 1 ml needleless syringe (Figure 2). The infiltrated plant were dried gently using the tissue paper and kept in the growth chamber for 3-6 h before collected and frozen in liquid nitrogen.

Figure 2. Infiltration of N. benthamiana using the needless syringe
- N. benthamiana plants were grown in a growth chamber at 19-22 °C under long-day conditions (14-h-light/10-h-dark) and 70-80% relative humidity for 5-6 weeks as shown in Figure 1.
- Total RNA isolation
Total RNA was isolated from N. benthamiana leaves using E.Z.N.A.® Total RNA Kit I isolation kit according to the manufacturer’s instructions with proper modifications.- Harvest the N. benthamiana leaves at 3-6 h after PAMP treatment. Grind the frozen tissues into powder with a mortar and pestle in liquid nitrogen.
- Transfer the leaf powder into the 1.5 ml RNase-free tube with 700 μl TRK Lysis Buffer and 14 μl β-mercaptoethanol, vortex the samples for 30 sec, and maintain the sample at 4 °C for 5-10 min.
- Centrifuge the samples at 15,000 x g for 5-10 min at 4 °C, carefully transfer 600 μl supernatant into a new 1.5 ml RNase-free tube.
- Add equal volume (600 μl) 70% ethanol (see Recipes).
- Add the mixed liquid into the HiBind® RNA Mini Column (assembled 2 ml collection tube), centrifuge at 10,000 x g for 30 sec at 4 °C. Discard the filtrate and reuse the collection tube.
- Add 300 µl RNA Wash Buffer I to the HiBind® RNA Mini Column, centrifuge at 10,000 x g for 30 sec at 4 °C. Discard the filtrate and reuse the collection tube.
- Add 500 µl RNA Wash Buffer I to the HiBind® RNA Mini Column, centrifuge at 10,000 x g for 30 sec at 4 °C. Discard the filtrate and change a new 2 ml collection tube.
- Add 500 μl RNA Wash Buffer II, centrifuge at 10,000 x g for 30 sec at 4 °C. Discard the filtrate and reuse the collection tube.
- Repeat Step B8.
- Centrifuge at 10,000 x g for 2 min to completely dry the HiBind® RNA Mini Column.
- Put the HiBind® RNA Mini Column into a new 1.5 ml RNase-free tube, add 50 μl DNase/RNase-free ddH2O into Column, and let it sit at room temperature for 1 min. Centrifuge at 10,000 x g for 1 min at 4 °C to elute RNA from the HiBind® RNA Mini Column.
- Measure RNA concentration using NanodropTM 1000. Store the eluted RNA at -70 °C.
- First-strand cDNA synthesis
Total RNA was reverse transcribed using PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time).- Remove gDNA reaction
Reaction mixture:
Reaction program:5x gDNA Eraser Buffer 2 μl gDNA Eraser 1 μl Total RNA 700 ng (calculate volume) DNase/RNase-free ddH2O up to 10 μl 42 °C 2 min 4 °C ∞ - Reverse transcription reaction
Reaction mixture:
Reaction program:The reaction solution of Step C1 10 μl PrimeScriptTM RT Enzyme Mix I 1 μl RT Primer Mix 1 μl 5x PrimeScriptTM Buffer 2 (for Real Time) 4 μl DNase/RNase-free ddH2O up to 20 μl 37 °C 15 min 85 °C 5 sec 4 °C ∞ - Dilute the obtained cDNA 20 times with 180 µl RNase-free H2O.
- Remove gDNA reaction
- Quantitative Real-time PCR (qRT-PCR)
qRT-PCR was performed using TB GreenTM Premix Ex TaqTM Kit (Tli RNase H Plus):- qRT-PCR reaction mix:
2x TB GreenTM Premix Ex TaqTM (Tli RNase H Plus) 10 μl 50x ROX Reference Dye II 0.4 μl qRT-PCR Forward Primer 0.4 μl qRT-PCR Reverse Primer 0.4 μl The diluted first-strand cDNA 5 μl DNase/RNase-free ddH2O up to 20 μl - Add the qRT-PCR reaction mix into each wall of MicroAmpTM Optical 96-Well Reaction Plate with Barcode with three replicates for each sample. Seal the plate using MicroAmpTM Optical Adhesive Film, and centrifuge the plate at room temperature for 3 min using the MPS 1000 Mini PCR Plate Spinner.
- qRT-PCR program:
Stage 1: Pre-denaturation at 95 °C for 30 sec Stage 2: PCR amplification (Reps: 40) 95 °C 5 sec 60 °C 34 sec Stage 3: Melting Curve 95 °C 15 sec 60 °C 1 min 95 °C 15 sec
- qRT-PCR reaction mix:
Data analysis
Ct (cycle threshold) values were collected and exported from the Sequence Detection Software v1.4.0. Relative expression of marker genes in each sample was normalized against the internal control (Endo), such as the housekeeping genes NbActin or NbEF1α, and was calculated using the formula: 2-ΔCT = 2-(Cttarget - CtEndo) calculated as ΔOD1-ΔOD2.
The relative expression level of marker genes in PAMP-treated samples to the control-treated sample was calculated using the following formula: 2-ΔΔCT = 2-(ΔCTPAMP - ΔCTcontrol) (Figure 3).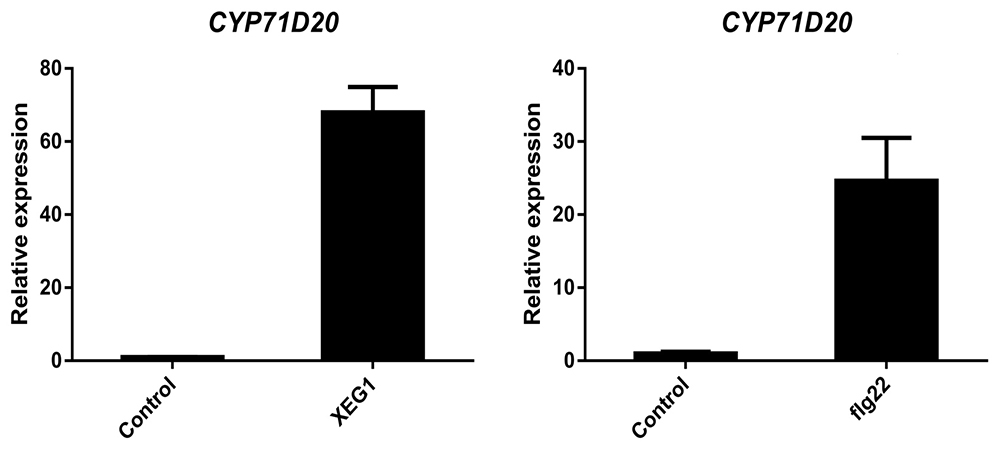
Figure 3. Transcription level changes of PTI marker gene CYP71D20 upon XEG1or flg22 treatment. Transcript levels were quantified by qRT-PCR and normalized to internal control NbEF-1α. Bars represent the mean fold changes (± S.E.M.) of the XEG1-treated or flg22-treated leaves relative to the value in control-treated leaves, which was set as 1.
Notes
- It is important to use plants of similar size. Leaves of different plants should be collected at the same position with similar size.
- The negative control should be included to measure the background level.
- Cross-contamination of the PAMPs and negative control should be avoided during infiltration.
- For each experiment, at least three replicates should be included for statistical analysis.
- Different PAMPs may induce the defense-related marker gene expression at different time points.
- The qRT-PCR program applies to Applied Biosystems 7500 fast Real-Time PCR System.
Recipes
- 1 mM flg22 stock solution
2.27 mg of flg22 (QRLSTGSRINSAKDDAAGLQIA) (Molecular Weight: 2272.52) was dissolved in 1 ml ddH2O, and store at -20 °C. The stock was diluted to a final concentration of 200 nM - 1 mM XEG1 stock solution
2.73 mg of XEG1 was dissolved in 1 ml ddH2O, and store at -20 °C. The stock was diluted to a final concentration of 200 nM - 70% ethanol
Add 30 ml DNase/RNase-free ddH2O to 70 ml of absolute ethanol
Acknowledgments
This protocol was partially modified from the previously published paper by Wang et al. (2018). This work was supported by the China National Science Funds to Yuanchao Wang for Innovative Research Groups (Grant No. 31721004) and the funds to Yan Wang (Grant No. 31501622 and KJQN201663).
Competing interests
The authors declare that they have no conflicts of interest or competing interests.
References
- Couto, D. and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16(9): 537-552.
- Felix, G., Duran, J. D., Volko, S. and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265-276.
- Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., Schroeder, J. I., Peck, S. C. and Rathjen, J. P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104(29): 12217-12222.
- Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., Dong, S., Zhang, Z., Dou, D., Zheng, X., Tyler, B. M. and Wang, Y. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27(7): 2057-2072.
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T. and Jones, J. D. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135(2): 1113-1128.
- Segonzac, C., Feike, D., Gimenez-Ibanez, S., Hann, D. R., Zipfel, C. and Rathjen, J. P. (2011). Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol 156(2): 687-699.
- Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., Ye, W., Lin, Y., Shao, Y., Dong, S., Tyler, B. M. and Wang, Y. (2018). Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun 9(1): 594.
- Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T. and Felix, G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125(4): 749-760.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Liu, F., Xu, Y., Wang, Y. and Wang, Y. (2018). Real-time PCR Analysis of PAMP-induced Marker Gene Expression in Nicotiana benthamiana. Bio-protocol 8(19): e3031. DOI: 10.21769/BioProtoc.3031.
Category
Plant Science > Plant molecular biology > RNA > Transcription
Plant Science > Plant physiology > Biotic stress
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










