- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Ni-NTA-Agarose Purification of Recombinant Hepatitis C Virus E2 Ectodomain Produced in a Baculovirus Expression System
(*contributed equally to this work) Published: Vol 8, Iss 19, Oct 5, 2018 DOI: 10.21769/BioProtoc.3030 Views: 6501
Reviewed by: David PaulJan-Ulrik DahlSteven James Burgess

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Production and Crystallization of Nanobodies in Complex with the Receptor Binding Domain of the SARS-CoV-2 Spike Protein
Audrey Le Bas [...] Raymond J. Owens
May 5, 2022 3704 Views

Purification of Crimean Congo Hemorrhagic Fever Virus (CCHFV) Nucleocapsid Protein Using Detergent Gradient and Free Thawing
Austin Royster and Sheema Mir
Aug 5, 2022 1966 Views

Carbamoyltransferase Enzyme Assay: In vitro Modification of 5-hydroxymethylcytosine (5hmC) to 5-carbamoyloxymethylcytosine (5cmC)
Weiwei Yang [...] Laurence Ettwiller
Sep 5, 2022 1986 Views
Abstract
In this protocol, we describe the production and purification of the ectodomain of the E2661 envelope protein (amino acids 384-661) of the Hepatitis C virus, which plays a fundamental role in the entry of the virus into the host cell. This protein has been expressed in both prokaryotic and eukaryotic systems but in small quantities or without native protein characteristics. In our case, we use the Baculovirus expression system in insect cells. E2661 is secreted into the extracellular medium and purified by means of affinity chromatography a Ni-NTA-column because the protein has a tag of six histidines at its amino terminal end. The purified protein possesses a native-like conformation and it is produced in large quantities, around 5-6 mg per liter.
Keywords: Hepatitis C virusBackground
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide (Major et al., 2001; Alter, 2006). At this moment, there is no vaccine for HCV and antivirals are used to treat the HCV infection (Imran et al., 2014). However, treatments are expensive and not 100% effective (Kohli et al., 2014). The HCV envelope glycoprotein E2 is responsible for the interaction with cellular receptors, thus it is a major candidate to study the first steps of the infective cycle of the virus. Previous expression systems produce low levels of heterogeneous protein due to glycosylation and aggregation, and it is difficult to distinguish between molecules that undergo productive and non-productive folding (Flint et al., 2000). In this protocol, we describe the production of the recombinant ectodomain of E2 tagged with a 6xHis extension at N-terminal end of the protein in a baculovirus/insect cell system. The gp67 signal peptide fused to the E2 ectodomain mediates the forced secretion of the recombinant protein. The protein is secreted to the cell supernatant and purified by means of affinity chromatography with a Ni-NTA-Agarose column. The yield of the process was 5-6 mg of protein per liter of media. This protein possesses a native-like conformation as determined by different spectroscopic techniques such as circular dichroism or fluorescence spectroscopy, as well as by its recognition in an enzyme immunoassay by a conformation specific antibody (Rodriguez-Rodriguez et al., 2009). The use of this independent folding domain that is able to acquire its proper folding in absence of the E1 glycoprotein, may contribute to shed light on the biology of HCV (three-dimensional or secondary structure of the protein and its role in the fusion of the HCV virus and the host cell membranes). Also, it could also be used as a vaccine in the prevention of HCV infection.
Materials and Reagents
- Pipette tips 200 μl (Sigma-Aldrich, catalog number: P5161 )
- Tissue culture flasks F75 (75 cm2 surface area) (TPP Techno Plastic Products, catalog number: 90075 )
- Tissue culture flasks F150 (150 cm2 surface area) (TPP Techno Plastic Products, catalog number: 90150 )
- Cell culture flasks F25 (25 cm2 surface area) (Corning, catalog number: 3055 )
- Tissue culture dish 35 mm (SARSTEDT, catalog number: 83.3900 )
- Sterile tube 50 ml (SARSTEDT, catalog number: 62.547.254 )
- Sterile tube 15 ml (Fisher Scientific, catalog number: 05-539-12 )
- Serological pipette 10 ml (SARSTEDT, catalog number: 86.1254.001 )
- Dialysis membrane Spectra/Por® 6 (VWR, Spectrum, catalog number: 734-0646 )
- Insect cell line Spodoptera frugiperda (Sf9) (Oxford Expression Technologies, catalog number: 100201 )
- Insect cell line Trichopulsia ni (High Five) (Tni) (generously donated by PhD J. Pérez-Gil, Dpt. Biochemistry and Molecular Biology, Faculty of Biology, University Complutense of Madrid, Madrid, Spain)
- Baculovirus transfer vector pAcGP37A (BD, BD PharmingenTM, catalog number: 21220P )
- Recombinant transfer vector pAcGP67A-E2661, obtained according to the procedure described in Rodriguez-Rodriguez et al., 2009)
- FlashBAC GOLD kit (Oxford Expression Technologies, catalog number: 100201 ) composed of:
- flashBACTM DNA
- Positive control transfer plasmid DNA (expressing lacZ)
- Baculofectin II
- Insect-XPRESSTM Protein-free Insect Cell Medium with L-glutamine (Lonza, catalog number: 12-730Q )
- Neubauer's counting camera (VWR, MARIENFELD, catalog number: 631-0696 )
- Gentamicin (Sigma-Aldrich, catalog number: G1264 )
- TC100 Insect Medium (Lonza, catalog number: BE02-011F )
- Ni2+-nitrilotriacetic acid agarose (Ni-NTA agarose) resin (QIAGEN, catalog number: 30230 )
- Tris-Base (Sigma-Aldrich, catalog number: T1503 )
- NaCl (Merck, catalog number: 106404 )
- Imidazole (Sigma-Aldrich, catalog number: 56750 )
- EDTA (Sigma-Aldrich, catalog number: 03695 )
- Dodecyl sulfate sodium salt (Merck, catalog number: 1.13760.1000 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Glycine (Sigma-Aldrich, catalog number: G8898-1KG )
- Bromophenol blue (United States Biological, catalog number: 12370 )
- 2-mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Acrylamide (Sigma-Aldrich, catalog number: A8887-500G )
- N,N′-Methylenebis(acrylamide) (Sigma-Aldrich, catalog number: M7279-250G )
- Ammonium persulphate (Sigma-Aldrich, catalog number: A3678-100G )
- N,N,N',N',N'-tetramethylethylenediamine (TEMED) (Bio-Rad Laboratories, catalog number: 1610801 )
- Coomassie Brilliant Blue R-250 (Sigma-Aldrich, catalog number: B8647 )
- Methanol (Sigma-Aldrich, catalog number: 322415-1L )
- Acetic acid (Merck, catalog number: 1000063 )
- 3x Protein electrophoresis application buffer (see Recipes)
- Running electrophoresis buffer (see Recipes)
- Blue staining solution (see Recipes)
- Bleaching solution (see Recipes)
- Separating gel (see Recipes)
- Concentrating gel (see Recipes)
Equipment
- Pipettes
- -80 °C freezer
- CO2 Water-jacketed Incubator (NuAire, model: NU-2700 IR Autoflow )
- Laminar flow chamber (Azbil Telstar, model: BV-100 )
- Laboratory centrifuge (MPW MED. INSTRUMENTS, model: MPW-223e )
- Upright microscope (Nikon Instruments, IZASA, model: H550S )
- 5 ml plastic syringe
- Mini-PROTEAN® Tetra Electrophoresis System (Bio-Rad Laboratories, catalog number: 165-8001 )
- Plastic bucket (Labotienda, catalog number: BTL006 )
- UV-Spectrophotometer (Shimadzu, model: UV-1800 )
- 15 L Plastic bucket for dialysis
Procedure
- Generation of recombinant baculoviruses by cotransfection
- Seed the 35 mm tissue culture dishes with 1.5 x 106 Sf9 cells in 2 ml of Insect-XPRESSTM medium with gentamicin at a final concentration of 10 μg/ml to form a sub-confluent monolayer. The number of insect cells is determined by using a Neubauer’s camera.
- Incubate for 1 h at 27 °C in the CO2 incubator to allow cell attachment to the plate.
- Add the following reagents to prepare the co-transfection mix of DNA and transfection reagent in a 1.5 ml polystyrene tube:
- 100 μl of serum-free medium (TC100)
- 100 ng of virus DNA from the flashBAC kit
- 500 ng of transfer plasmid (pAcGP67A-E2661 recombinant plasmid or lacZ positive control transfer vector from the flashBAC kit)
- 1.2 μl of baculoFECTIN II from the flashBAC kit
- Wash the cell monolayers twice by pouring 5 ml of TC100 medium without serum onto monolayer, shake gently and decant into waste. Add 1 ml of TC100 medium without serum to each 35 mm dish. If the cells are maintained in serum-free medium, it is not necessary to wash the monolayer. In this case, remove and discard 1 ml of medium from the 35 mm dishes.
- Add the transfection mix (111.2 μl) to the 35 mm dish.
- Incubate overnight at 27 °C in the CO2 Incubator without shaking.
- Add 1 ml of Insect-XPRESSTM medium with gentamicin at 10 μg/ml to the 35 mm dish.
- Incubate at 27 °C for 4 days in the CO2 Incubator without shaking.
- Harvest the culture medium that contains the recombinant virus by decanting the medium into a 15 ml sterile container. Centrifuge in the laboratory centrifuge for 15 min at 325 x g. Collect the supernatant and store in the dark in a refrigerator at 4 °C. For a long-term storage, save the container at -80 °C.
- Amplification of recombinant viruses
Note: Once the purified recombinant baculovirus is available, it must be amplified in order to obtain recombinant protein.- Seed 2 x 106 Tni insect cells per 25 cm2 of surface area and incubate for 1 h at 27 °C.
- Add 1 ml of the supernatant containing the recombinant viruses for every 25 cm2 of surface area (Flask F25, 1 ml; Flask F75, 3 ml; Flask F150, 6 ml).
- Incubate for one hour at 27 °C in the CO2 incubator with very slow agitation (rock the flask every 10 min) to maintain the hydration of the cell monolayer with the minimum volume of inoculum.
- Add the relevant volume of Insect-XPRESSTM medium with gentamicin at 10 μg/ml to the flask (Flask F25, until 5 ml; Flask F75, until 10 ml; Flask F150, until 30 ml).
- Incubate at 27 °C for 5 days in the CO2 Incubator without agitation.
- Collect the supernatant in a 15 or 50 ml sterile tube in order to recover the amplified recombinant virus and centrifuge in the laboratory centrifuge for 15 min at 325 x g.
- Use the supernatant for a second round of amplification. Transfer the supernatant to a separate sterile tube and repeat Steps B2 to B6. Usually, two or three rounds of amplification are sufficient for large-scale infection to allow expression of the recombinant protein of interest.
- Production and Purification of HCV E2661 protein
- Prepare 10 flasks of 150 cm2 surface area with 8-10 x 106 Tni insect cells in 20 ml medium with gentamicin at 10 mg/ml and incubate for one hour at 27 °C in the CO2 incubator without agitation.
- Remove the medium and add 6 ml of supernatant containing the recombinant E2661 baculovirus per flask.
- Incubate for one hour at 27 °C in the CO2 incubator with very slow agitation (rock the flask every 10 min) to maintain the hydration of the cell monolayer with the minimum volume of inoculum.
- Add 24 ml of Insect-XPRESSTM medium with gentamicin at 10 μg/ml per flask.
- Incubate at 27 °C for 5 days.
- Collect the supernatants and centrifuge for 15 min at 325 x g.
- Dialyze the supernatant (300 ml, 10 flasks of 30 ml) against 20 L (10 L, 2x) of 50 mM Tris-HCl, pH 8.0, 300 mM NaCl buffer in a 15 L plastic bucket.
- Load the supernatant onto a 2 ml Ni-NTA-Agarose column previously equilibrated with 50 mM Tris-HCl, pH 8.0, 300 mM NaCl buffer.
- Wash the column by adding 20-40 ml of 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM Imidazole buffer to the top, let the solution flow into a beaker to collect the waste. Wash until optical density at 280 nm is below 0.03. The adsorption at 280 nm is determined by using an UV-spectrophotometer.
- Wash the column by adding 10-30 ml 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 30 mM Imidazole until optical density at 280 nm is below 0.03.
- Elute the E2661 protein by adding 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 200 mM Imidazole. Collect fractions of 1-1.2 ml. either in 1.5 ml Eppendorf or in glass tubes. Usually 10-12 tubes are enough for elution; thus, manual collection with the operator changing the tube after 1 ml is recommended.
- The protein is detected by measuring the optical density at 280 nm of each fraction in the UV-spectrophotometer.
- Check the purification steps of the recombinant E2661 protein by SDS-PAGE. In order to check the purity of the protein, load 20 μl of sample per well from each 200 mM Imidazole fraction. After Coomassie blue staining, the purity of recombinant E2661 is tested by the appearance of a unique, isolated band at 48 kDa.
- Collect the fractions containing significant amounts (optical density at 280 nm higher than 0.3) of pure E2661 protein in a single fraction.
- Dialyze the E2661 protein against 50 mM Tris-HCl, pH 8.0, 300 mM NaCl buffer.
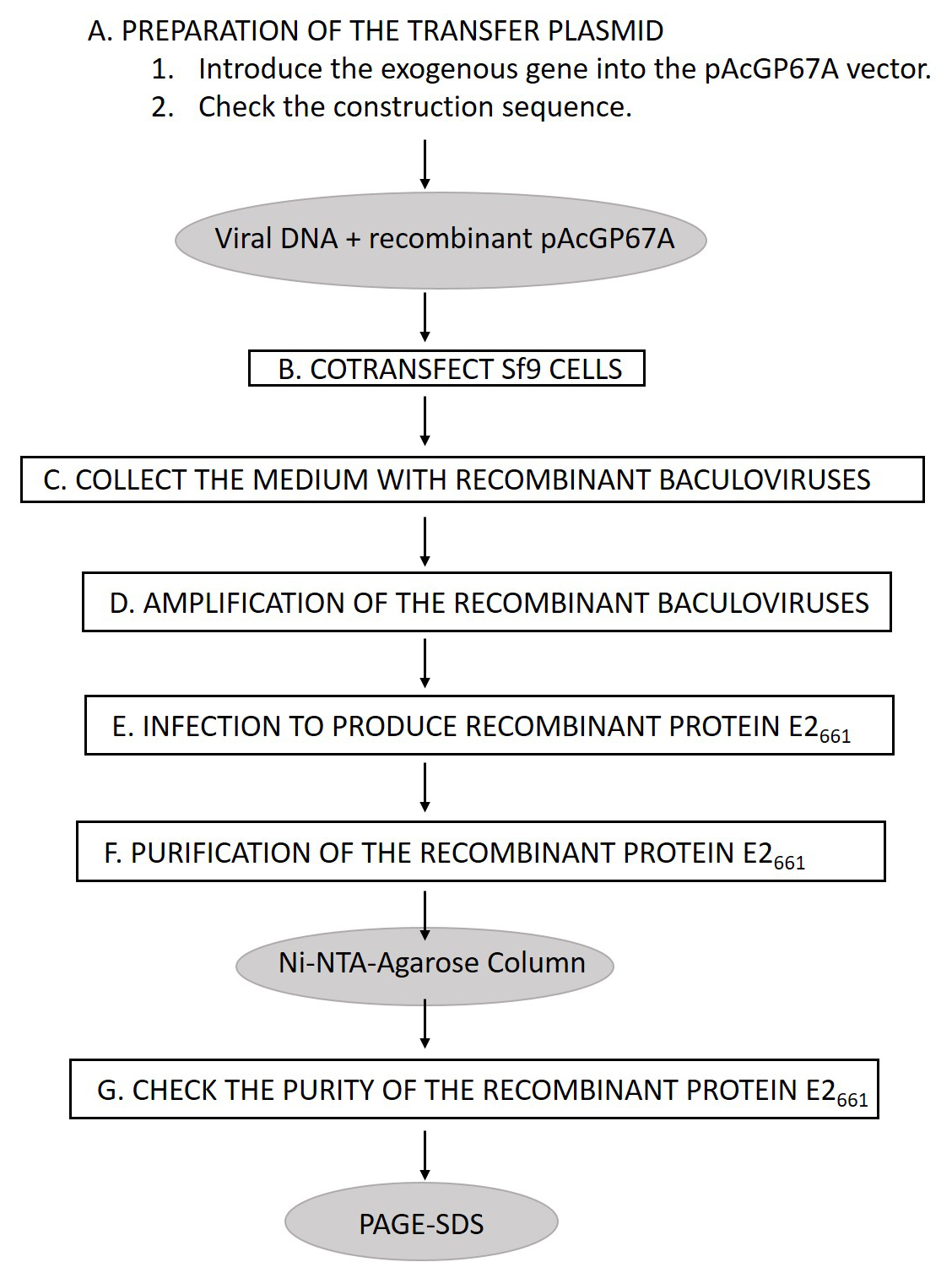
Figure 1. Scheme to produce the recombinant protein E2661 - SDS-PAGE (Protein electrophoresis)
Note: A more detailed protocol to carry out SDS-PAGE is described in He (2011) and Yan (2011).- Prepare the electrophoresis system.
- Prepare the separating gel (see Recipe 5).
- Mix with a Pasteur pipette and deposite into the gel stand.
- Prepare the concentrating gel (see Recipe 6).
- Mix with a Pasteur pipette and deposit on top of the separating gel.
- Add 10 μl of 3x Protein electrophoresis application buffer (see Recipe 1) to 20 μl of sample.
- Heat at 95 °C for 5 min.
- Load into the concentrating gel.
- Develop the electrophoresis in the Running electrophoresis buffer (see Recipe 2) at room temperature at 25 mA per gel until the marker reached the end of the gel.
- Incubate the gel in a Coomassie blue staining solution (see Recipe 3) at room temperature for 10 min.
- Incubate the gel in the bleaching solution (see Recipe 4) to detect the proteins.
Data analysis
The results of a standard purification process, corresponding to a PAGE-SDS gel, are shown in Figure 2. Lanes 2, 3 and 4 show washing steps with 10 mM (lane 2) and 30 mM (lanes 3 and 4) Imidazole buffer, where some contaminant proteins weakly attached to the Ni-NTA-agarose column are eliminated. As can be seen, the protein E2661 remains in the column and is only eluted when the 200 mM Imidazole buffer is used (Figure 2, lanes 5 and 6). As shown in the gel, the recombinant protein is pure.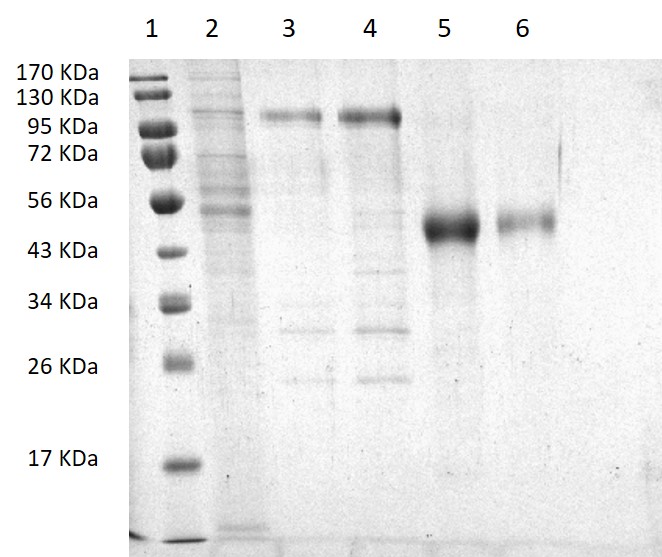
Figure 2. SDS-PAGE of the purification steps of the recombinant protein E2661. (1) Molecular weights ladder; (2) Wash with 10 mM Imidazole buffer; (3, 4) Wash with 30 mM Imidazole buffer; (5, 6) E2661 protein eluted with 200 mM Imidazole buffer.
Recipes
- 3x Protein electrophoresis application buffer
Tris 150 mM, pH 7.6
EDTA 6 mM
3% (w/v) SDS
30% (v/v) Glycerol
0.06% (w/v) Bromophenol blue
15% (v/v) β-mercaptoethanol - Running electrophoresis buffer
Tris 0.025 M, pH 8.3
Glycine 0.192 M with 0.1% SDS - Blue staining solution
0.3% (w/v) Coomassie Brilliant Blue R-250
45% (v/v) Methanol
10% (v/v) Acetic acid - Bleaching solution
7.5% (v/v) acetic acid
20% (v/v) methanol - Separating gel
650 μl H2O
1.82 ml Tris 1 M, pH 8.8
2.5 ml acrylamide-bisacrylamide at 30%
55 μl SDS 10%
12 μl of 0.075% TEMED
14 μl 0.02% (w/v) ammonium persulfate - Concentrating gel
2.0 ml acrylamide-bisacrylamide at 4%
55 μl SDS 10%
8 μl of 0.075% TEMED
8 μl of 0.02% (w/v) ammonium persulfate
Acknowledgments
This work was supported by grant BFU 2006-13033 from the Ministerio de Educación y Ciencia, Spain and the research project SANTANDER/COMPLUTENSE PR26/16-20271.
Competing interests
Authors declare that there are any conflicts of interest or competing interests.
References
- Alter, H. (2006). Viral hepatitis. Hepatology 43(2 Suppl 1): S230-234.
- Flint, M., Dubuisson, J., Maidens, C., Harrop, R., Guile, G. R., Borrow, P. and McKeating, J. A. (2000). Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J Virol 74(2): 702-709.
- He, F. (2011). Laemmli-SDS-PAGE. Bio-protocol Bio101: e80.
- Imran, M., Manzoor, S., Khattak, N. M., Khalid, M., Ahmed, Q. L., Parvaiz, F., Tariq, M., Ashraf, J., Ashraf, W., Azam, S. and Ashraf, M. (2014). Current and future therapies for hepatitis C virus infection: from viral proteins to host targets. Arch Virol 159(5): 831-846.
- Kohli, A., Shaffer, A., Sherman, A. and Kottilil, S. (2014). Treatment of hepatitis C: a systematic review. JAMA 312(6): 631-640.
- Major, M. E., Rehermann, B. and Feinstone, M. (2001). Hepatitis C Viruses. In: Knipe, D. M. and Howley, P. M. (Eds.). Fields Virology. Lippincott Williams & Wilkins, 1.
- Rodriguez-Rodriguez, M., Tello, D., Yelamos, B., Gomez-Gutierrez, J., Pacheco, B., Ortega, S., Serrano, A. G., Peterson, D. L. and Gavilanes, F. (2009). Structural properties of the ectodomain of hepatitis C virus E2 envelope protein. Virus Res 139(1): 91-99.
- Yan, Q. (2011). Rapid coomassie protein SDS-gel staining. Bio-protocol Bio101: e83.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gómez-Gutiérrez, J., Rodríguez-Rodríguez, M., Gavilanes, F. and Yélamos, B. (2018). Expression and Ni-NTA-Agarose Purification of Recombinant Hepatitis C Virus E2 Ectodomain Produced in a Baculovirus Expression System. Bio-protocol 8(19): e3030. DOI: 10.21769/BioProtoc.3030.
Category
Biochemistry > Protein > Expression
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








