- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Plant Assays for Quantifying Ralstonia solanacearum Virulence
Published: Vol 8, Iss 18, Sep 20, 2018 DOI: 10.21769/BioProtoc.3028 Views: 9028
Reviewed by: Modesto Redrejo-RodriguezDavid NormanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A β-glucuronidase (GUS) Based Bacterial Competition Assay to Assess Fine Differences in Fitness during Plant Infection
Julien S. Luneau [...] Alice Boulanger
Jul 5, 2022 3236 Views

Measurements of Root Colonized Bacteria Species
Vílchez Juan Ignacio [...] Huiming Zhang
Apr 5, 2021 6351 Views

Tomato Stem Injection for the Precise Assessment of Ralstonia solanacearum Fitness in Planta
Yaru Wang [...] Alberto P. Macho
Aug 20, 2021 3745 Views
Abstract
Virulence assays are powerful tools to study microbial pathogenesis in vivo. Good assays track disease development and, coupled with targeted mutagenesis, can identify pathogen virulence factors. Disease development in plants is extremely sensitive to environmental factors such as temperature, atmospheric humidity, and soil water level, so it can be challenging to standardize conditions to achieve consistent results. Here, we present optimized and validated experimental conditions and analysis methods for nine assays that measure specific aspects of virulence in the phytopathogenic bacterium Ralstonia solanacearum, using tomato as the model host plant.
Keywords: Virulence assayBackground
Ralstonia solanacearum is a soil-borne bacterium that causes bacterial wilt in a vast range of plants and continues to infect new hosts across the globe (Hayward, 1991; Elphinstone, 2005; Wicker et al., 2007; Genin, 2010; Weibel et al., 2016). As a result, R. solanacearum is among the most intensively studied phytopathogenic bacteria (Mansfield et al., 2012).
R. solanacearum can persist in soil or water reservoirs for long periods (Alvarez et al., 2008), and in the presence of a suitable host, it can enter the plant through wounds or lateral root emergence points (Denny, 2007). Thereafter it colonizes the water-transporting plant xylem vessels and thrives there. Massive production of exopolymeric substances (EPS) likely contributes to the clogging of the xylem channels, leading to blockage of water transport, followed by symptoms like wilting leaves, stunted growth, stem discoloration, and death. Molecular genetic studies revealed a consortium of many virulence factors that are required for pathogenesis and fitness in planta (Genin and Denny, 2012; Tran et al., 2016a and 2016b); recent in silico modeling (Peyraud et al., 2016) and in vivo transcriptomics and metabolomics (Jacobs et al., 2012; Khokhani et al., 2017; Lowe-Power et al., 2018) have further enhanced our understanding of how this bacterium switches from saprophytic to parasitic lifestyle.
To test hypotheses suggested by molecular data, researchers measure virulence on model host plants under controlled conditions. To be useful, such assays must be quantitative, biologically relevant, and replicable. We have developed or adapted several protocols to assess R. solanacearum interactions with tomato, a natural host and economically important crop plant. A naturalistic soil soak assay replicates many aspects of the infection process that occurs in the field. This assay quantifies the defects of mutant strains lacking virulence factors involved in the early phases of the disease such as sensing, invading, and colonizing host roots. For example, mutants lacking chemotaxis, swimming motility, extracellular plant cell wall-degrading enzymes, and type II and III secretion systems are all impaired in virulence following soil soak inoculation.
A petiole inoculation disease assay that introduces the pathogen directly into stem xylem vessels can identify traits that contribute to pathogen success in xylem vessels (Saile et al., 1997). Some mutants that are defective in virulence following soil soak inoculation have full wild-type virulence following cut-petiole inoculation into the stem; examples include mutants lacking motility and chemotaxis (Tans-Kersten et al., 2001; Yao and Allen, 2006). In another case, comparing results of these two assays revealed that extracellular DNA degradation, a trait initially thought to only play a role in interactions with host roots, was also critical for normal biofilm formation inside host xylem later in disease (Tran et al., 2016a and 2016b). When virulence traits are functionally redundant or make small contributions to pathogen success, neither soil soak nor petiole inoculation assays can reveal subtle differences between the wild-type and the target mutant strain (Macho et al., 2010). In those cases, we can use single or co-inoculation experiments to directly compare the growth of competing strains in planta (root and/or shoot colonization) and calculate their relative competitive fitness as a competitive index (CI). We also describe here the protocol to measure bacterial attachment to plant roots. Since R. solanacearum is a xylem-dwelling bacterium, it is important to understand how it affects and is affected by host plant xylem sap. Therefore, we provide a protocol for collection of xylem sap from healthy and infected tomato plants; this ex vivo sap can be used as a medium for bacterial growth curves or for metabolomic analyses.
Materials and Reagents
- Sterile conical flask 250 ml (Corning, PYREX®, catalog number: 4980-250 )
- Sterile 50 ml conical tubes (Stellar Scientific, catalog number: T50-100 )
- Flasks for preparing large volume of cultures (size depends on the experimental goals)
- Seedling tray (36-cell insert with holes) (J&P Park Acquisitions, Park Seed, catalog number: 96377 )
- Flat trays (greenhouse megastore, 11" W x 21.37" L x 2.44" D, CN-FL)
- 1.5 ml microcentrifuge tubes (Sigma-Aldrich, BRAND, catalog number: Z336769 )
- 8 cm wide pots (greenhouse megastore)
- Planting sticks (greenhouse megastore)
- Metal beads (2.4 mm) (OMNI, catalog number: 19-640 )
- Bead-beater tubes (USA Scientific, catalog number: 1420-9300 )
- Gosselin Square Polystyrene Petri Plate with 4 vents, 120 x 120 x 15.8 mm, Sterile (Corning, catalog number: BP124-05 )
- MicroporeTM Surgical Paper Tape (1 inch size) (3M, catalog number: 1530-1 )
- Aluminum foil (W.W. Grainger, catalog number: 16W479 )
- Paper towel (Singlefold Paper Towel, 9.1 x 10.25) (Cascades Pro, catalog number: H165 )
- WhatmanTM paper (Grade 1 Qualitative Filter Paper) (GE Healthcare, Whatman)
- Petri dish (Corning, Falcon®, catalog number: 351029 )
- 0.22-μm sterile filter (Merck, catalog number: SLGP033RS )
- 10 ml pipette (Disposable Polystyrene Serological Pipettes) (Corning, Falcon®, catalog number: 357551 )
- 1 ml syringe (New Sterile, Sealed, Tuberculin, Luer slip tube, No Needle, Disposable) (BD, catalog number: 9602 )
- 96-well half-area microplates (Corning, catalog number: 3697 )
- Soil mix, propagation mix, Sunshine® resilience silicon enriched, Re Plug and Seed Rsi (Sun Gro Horticulture, Lot-code: Q15322; SKU: 7263924)
Ingredients:
55-65% Canadian Sphagnum peat moss, vermiculite, dolomite lime, wetting agent;
0.0001% Silicon dioxide (SiO2) from calcium silicate to increase root growth - Ralstonia solanacearum strain from glycerol or water stock
- Bonny best wilt susceptible variety of Tomato seeds (Mountain valley seed) (stored at 4 °C)
- Sterile reverse osmolyzed water by Milli-Q system (SMQ)
- Agar (Fisher Scientific, catalog number: BP1423-2 )
- Bleach (Clorox Performance Bleach with CloroMax) (The Clorox Company, catalog number: 980042447 )
- 70% Ethanol (Diluting 100% Ethanol 200 proof) (Decon Labs, catalog number: 2716 )
- Glucose (Fisher Scientific, catalog number: D16-1 )
- Peptone (Fisher Scientific, catalog number: NC9931583 )
- Casamino acids (RPI, catalog number: C45000-5000.0 )
- Yeast extract (Fisher Scientific, catalog number: BP1422-2 )
- KOH (Fisher Scientific, catalog number: P250 10 )
- 1% 2,3,5-triphenyl tetrazolium chloride (TZC) (Sigma-Aldrich, catalog number: T8877 )
- KNO3 (Fisher Scientific, catalog number: P383 100 )
- KH2PO4 (Merck, Calbiochem, catalog number: 529568 )
- MgSO4 (MP biomedicals, catalog number: 150136 )
- Ca(NO3)2•4H2O (Fisher Scientific, catalog number: C109 )
- H3BO3 (Fisher Scientific, catalog number: A73 1 )
- MnCl2•4H2O (Sigma-Aldrich, catalog number: M8054 )
- ZnSO4•7H2O (Fisher Scientific, catalog number: Z68 )
- CuSO4•5H2O (VWR, BDH, catalog number: BDH9312 )
- (NH4)6Mo7O24 (Sigma-Aldrich, catalog number: A1343 )
- FeSO4•7H2O (Fisher Scientific, catalog number: I146 3 )
- Na2EDTA (Fisher Scientific, catalog number: S311 )
- Casamino acid-peptone-glucose (CPG) agar (see Recipes)
- CPG broth (see Recipes)
- Modified Hoagland's solution (see Recipes)
Note: All the chemicals were purchased from Sigma-Aldrich, Fisher Scientific, or other chemical companies.
Equipment
- P1000 pipette (Eppendorf, catalog number: 3120000062 )
- P200 pipette (Eppendorf, catalog number: 3120000054 )
- P10 pipette (Eppendorf, catalog number: 3120000020 )
- Forceps (VWR, catalog number: 470018-952)
Manufacturer: Dunrite Instruments, catalog number: 141001 . - Scalpel (Bard-Parker® SafeSwitchTM Reusable Scalpel Handle, Size 3 L) (Aspen Surgical, catalog number: ST-1013LNS )
- Sharp razor blade (Carbon Steel Razor Blades) (Azpack, catalog number: YSJ-762-Q )
- Incubator (6M Lab Incubator) (Precision Scientific, catalog number: 31487 )
- Benchtop Shaker (Thermo Fisher Scientific, model: MaxQTM 4000 )
- Growth chamber with the following conditions:
Light intensity: 300-500 μmol/m2•sec-1
12 h, light, 28 °C
12 h, dark, 28 °C
50-70% humidity
~500 ppm CO2 measured - Powerlyzer® 24 homogenizer (MO BiO Laboratories, catalog number: 13155 )
- Centrifuge (15 amp version) (Eppendorf, model: 5810 R )
- Centrifuge (Eppendorf, model: 5417 R )
- Spectrophotometer (UV/Vis) (Beckman Coulter, model: DU 730 )
- Vortex mixer (Vortex-Genie 2) (Scientific industries, catalog number: SI-0246 )
- Biosafety cabinet (The Baker, SterilGard®, model: SG403A-HE )
- Balance (Roche Diagnostics, model: 05942861001 )
- Autoclave (Vacuum Steam Sterilizer) (Getinge, model: 533LS-E )
Software
- PRISM Graphpad software
Procedure
This section includes subsections from Procedure A to Procedure I:
- Preparing the bacterial culture for soil soak (depending on the goal of the experiment, this subsection can be followed by Procedures C-F or C-E-G or C-G)
- Sowing and transplanting of tomato plants–this subsection describes how to grow tomato plants to ensure reproducible results for soil soak or petiole inoculation (C or D) which can be followed by measuring disease progress (F) or competition and bacterial colonization (E and/or G).
- Soil soak inoculation–pouring a dilute bacterial suspension into pots containing unwounded tomato plants, which closely resembles the natural root infection process. Infection by this method is stochastic. This procedure can be followed by Procedure E or F or G or H, or E-G.
- Petiole inoculation–this alternative inoculation procedure is less natural but allows on to determine if a particular gene contributes to R. solanacearum success in the host stem. Petiole inoculation also ensure synchronized infection. (This procedure can be used for Procedure E or F or G or H, or E-G.)
- Competition assay–to quantify relative competitive fitness of wild-type and mutant R. solanacearum, we measure bacterial colonization in stems (Procedure G) from 3 to 7 days post inoculation or later (depending on the experimental goals).
- Disease progress curve–this procedure is used to quantify bacterial wilt disease development following soil soak (Procedure C) or petiole inoculation (Procedure D).
- Bacterial colonization–this procedure is used to measure the ability of bacteria to colonize the roots (early stages of infection) or stem (later stages of infection). This method quantified R. solanacearum population size in planta following soil soak (Procedure C) or petiole inoculation (Procedure D) or competition assay (Procedure E).
- Xylem sap collection–ex vivo tomato xylem sap can be used for either metabolomic analyses or as a medium for bacterial growth. Generally plants are infected by naturalistic soil soak method (Procedure A-B-C).
- Root attachment assay–to quantify the ability of mutants vs. wild-type bacteria to attach to tomato seedling roots, one of the very first steps in R. solanacearum infection of plants.
- Preparing the bacterial culture for soil soak
- Streak out Ralstonia solanacearum from water stock or glycerol stock on a CPG + TZC plate and let it grow in a 28 °C static incubator for 48 to 72 h.
Note: Make sure the morphology of the colonies is of wild-type (WT) and not spontaneous phcA- like mutant (Khokhani et al., 2017). - Once you see the isolated colonies on the plate, pick one colony to inoculate 100 ml of CPG broth (with appropriate antibiotics) in a 250-ml sterile flask and culture overnight (18-20 h) in a shaker-incubator at 220 rpm, 28 °C.
- Centrifuge (6,900 x g, 5 min) the overnight culture in a 50 ml conical tube twice to pellet all 100 ml of cell culture. Discard the supernatant.
Note: When you decant the supernatant, some of the EPS tends to come off too and that is a good sign that the culture is not contaminated with spontaneous phcA like mutants that do not make wild-type level of EPS. - Resuspend the cell pellet in 20 ml of sterile MilliQ (SMQ) water using a P1000 pipette, then add the remaining 30 ml of SMQ water and mix well by vortexing.
- Measure the optical density (λ = 600 nm, in a spectrophotometer) of the overnight cell culture by diluting it 10-fold in SMQ by pipetting 100 μl of resuspended cell culture in 900 μl of SMQ water in a 1.5 ml Eppendorf tube.
Example: If the OD600 is 0.5 after diluting, the actual OD600 is 5.0. - Calculate the volume of original cell suspension required to generate the desired volume of bacterial suspension at OD600 = 0.1, which corresponds to about 108 CFU/ml.
For one biological replicate that has 10 technical replicates, we do the following math:
For soil-soak inoculation, we add 5 x 109 cells per pot (50 ml of 0.1 OD600 cell suspension). So overall, we need 10 x 50 ml = 500 ml of cell suspension. Therefore, for 500 ml of 0.1 OD600 culture, we need 10 ml of 5.0 OD600 resuspended cell culture and the remaining 490 ml of SMQ water.
Note: Make a little extra inoculum so that you do not run out of cells for the last pot due to pipetting error. Generally, three biological replicates, each containing 14-20 technical replicates (plants) per strain are preferred for statistical analysis. - Check the OD600 again to make sure it is around 0.1 and dilution plate to confirm final inoculum cell density.
Note: Please see the attached Excel sheet for calculation.
- Streak out Ralstonia solanacearum from water stock or glycerol stock on a CPG + TZC plate and let it grow in a 28 °C static incubator for 48 to 72 h.
- Sowing and transplanting tomato seeds
- Add dry potting mix into a seedling tray with a gentle press.
- Place 4-5 seeds of Bonny Best (bacterial wilt-susceptible variety) in each cell.
Note: We have determined that tomato cv. Moneymaker is also similarly susceptible to bacterial wilt. - Cover them with a thin (1-3 mm) layer of potting mix as shown in Figure 1.

Figure 1. Sowing the tomato seeds in multi-pot-tray
- Carefully water the soil until the soil is wet and not flooded.
- Incubate the trays in the growth chamber at 28 °C with 12 h light/12 h dark cycles.
Note: High light intensity in the growth chamber is essential for good disease development. - Water the soil without flooding the tray (water should not flow out of the tray). Try to water the plants every morning around the same time.
- Stop watering the seedling tray on the 13th day after planting so that the soil gets a little dry. This allows the soil to fall off seedlings easily with a little shaking and makes it easy to transfer the seedlings to bigger pots. If the soil is wet, many roots can break even with gentle shaking.
- On the 14th day (Figure 2), transfer the seedlings from the seedling tray to bigger pots (Figure 3) by shedding away soil carefully (not to break the roots as much as possible). To remove soil, gently tap root ball as soil falls away.

Figure 2. Seedling growth on day 14th of planting seeds
Figure 3. Transplantation pots with soil mix ready for transfer of 14-day old tomato seedlings - Fill 8-cm diameter plastic pots with potting mix. It is important to not compress the soil to avoid creating an anaerobic condition in the pots.
- Water the soil until it is wet but not flooded and use your index finger or a Sharpie to make a hole in the center of the pot.
- Put seedlings onto the pots and when all pots have seedlings, put each seedling into a hole and gently press the soil around the base of the seedling so it is well-supported and will not fall over after being watered.
- Let the tomato plants grow in the bigger pots for another week till they are 21 days old. The first true leaf should be fully expanded and the second true leaf should be opening.
- Soil soak inoculation assay
The following protocol has been used previously (Huang and Allen, 2000; Tans-Kersten et al., 2001; Colburn-Clifford et al., 2010).- Stop watering the plants one or two days before inoculation, depending on the soil humidity. This will allow the soil to soak up all the bacterial culture poured into it. Be careful not to over dry, as R. solanacearum survival decreases in dry conditions.
- On the day before the inoculation of tomato plants, start an overnight culture of Ralstonia solanacearum as mentioned above in Step A1.
- On the day of inoculation follow Steps A2 to A7.
- Once you prepare the bacterial cultures, perform dilution plating of the inoculum and incubate the CPG plates for about 48 h to count the colonies. This will confirm that the OD600 was around 0.1 and that the bacterial cells were not contaminated or killed during the preparation.
- To blind-rate plants (preferred procedure), ask a colleague to code your inoculum flasks (e.g., with colored tape) and remove the identifying strain label from the flask. Colleague can keep the strain code list until you are ready to analyze data after the experiment is complete.
- Measure 50 ml of bacterial suspension using 50 ml conical tubes and pour this into each pot. You can do the same thing for all the target strains you aim to test for virulence, but use only one tube for each bacterial strain. Make sure to put planting sticks into the pots to label the plants for future disease rating (Figure 4). Do not wound plants.

Figure 4. Plant pots with planting sticks ready for inoculation - Check soil moisture within a few hours. For consistent results, the soil should not be desiccated for several days following inoculation. Dry soil will reduce bacterial viability and delay disease onset.
- After inoculation, water the plants following day with Hoagland’s solution. For the remaining 13 days use tap water to water the plants.
- Petiole inoculation assay
The following protocol has been used in (Brown and Allen, 2004; Dalsing and Allen, 2014).
Note: Follow all the steps mentioned in Procedure B and water the plants for all 21 days of plant growth.- On the day before inoculation, start 5 ml overnight CPG cultures of R. solanacearum.
- On the day of inoculation, water the plants (now 21-day old) first in the morning and then proceed to inoculation.
- Next day, spin down 1 ml of the overnight culture, and resuspend the pellet in 1 ml of SMQ water.
- Measure the OD600 and dilute the bacterial suspension in SMQ H2O to make 2 ml of OD600 = 0.1 bacterial cell suspension.
- Before moving forward, confirm the OD600 by spectrophotometer and dilution plating as described in Step A6.
- Dilute the 0.1 OD cell suspension to OD600 = 0.001.
- To infect the plant, cut off the first true leaf near the base of the petiole with a sharp razor blade. The petiole should be cut diagonally, parallel to the floor, to create a horizontal surface where a droplet of bacteria can be applied without running down the stem (Figure 5).
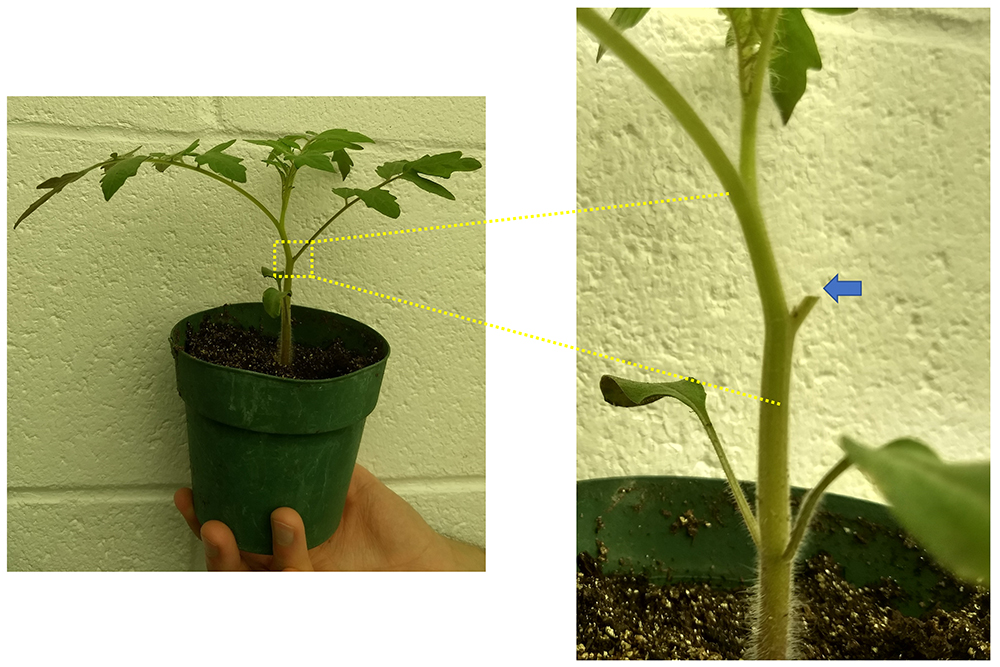
Figure 5. Petiole inoculation of 21-day old tomato plant. The blue arrow indicates the horizontal stump of cut petiole where 2 μl of bacterial cell suspension is placed. - Use a 10 ml pipette to carefully place 2 μl of OD600 = 0.001 cell suspension (equivalent to 2,000 cells) on the cut petiole surface. One can change the number of cells depending on the experimental goals. The droplet will be quickly absorbed by the plant, indicating that the bacteria were pulled into the xylem.
- You can cut all the petioles of plants in one tray and then apply the bacteria to each plant. For example, 10 plants at a time.
- Competition assay to measure relative competitive fitness of bacterial strains in planta
The following protocol has been used in (Yao and Allen, 2006; Lowe et al., 2015).- To measure the competitive fitness of the target strain relative to another strain (usually the wild-type) (Yao and Allen, 2006), each strain must carry a different antibiotic resistance. For example if the wild-type strain is kanamycin-resistant and the target mutant strain is gentamicin-resistant.
Note: To assure that antibiotic resistance marker gene does not add a fitness cost to the marked strain, compare the in planta growth of marked strains to growth of the unmarked parent strain. - Prepare fresh plates of the marked strains on CPG plates supplemented with the required antibiotics for overnight culture.
- Grow the plants as mentioned in Procedures B and C.
- To inoculate the plants (on 21st day of plant growth), separately prepare 0.1 OD600 cultures of the two competing strains. Check the OD600 using spectrophotometer.
- Combine the two strains in a 1:1 ratio of bacterial population. Again, dilution plate to check the actual number of cells.
- Add 50 ml of this mixed bacterial suspension to each pot for naturalistic soil soak competition, or 2 μl of the same suspension to the cut petiole for petiole inoculation competition.
- To check the root or stem colonization, sample the plants over 3 to 7 days post inoculation. From here, follow Procedure G.
- To measure the competitive fitness of the target strain relative to another strain (usually the wild-type) (Yao and Allen, 2006), each strain must carry a different antibiotic resistance. For example if the wild-type strain is kanamycin-resistant and the target mutant strain is gentamicin-resistant.
- Measuring Disease Progress, and creating a disease progress curve
- Before rating plants, water the plants first, wait for 1 h, and then record the disease by looking for wilted leaves. We record the disease on the scale from 0 to 4. If no leaves are wilted, the plant is scored as Disease Index (DI) = 0; if 24% or less of the leaf area is wilted we score that as DI = 1; if 25-49% is wilted, DI = 2; 50-74%, DI = 3; and 75 -100%, DI = 4.
- We rate the disease symptoms until 14 days post inoculation. Wilt symptoms normally begin to appear 3-4 days post petiole inoculation or 4-5 days post soil-soak inoculation.
- Measuring bacterial colonization of tomato roots and stems
The protocol for root colonization has been used in (Lowe et al., 2015), and stem colonization has been used in (Dalsing et al., 2015; Lowe-Power et al., 2016).- Sample collection depends on the experimental goal. Please refer to published articles where the sampling time is guided by the objective of the study.
- For root colonization, take the plants out of pots and gently shake off as much soil as possible.
- Put the plant in a container filled with water and let the roots soak and wash gently until all the soil comes off. Repeat the procedure for all the plants per treatment. (Figures 6A-6C)
- Wash roots for 15 sec in bleach, then successively for 15 sec in 2 fresh water baths and dry for 30 sec on a tissue (Figures 6D-6F).
- Cut off the roots at the crown junction (Figure 6G).
- Collect the roots and cut them into small pieces with a razor blade to ensure that each sample contains roots from every region (Figure 6H).
- Place up to 300 mg of the cut roots into a prepared bead-beater tube containing 4 small metal beads and 700 μl of SMQ water.
- For stem colonization, use a razor blade to harvest 1 cm of stem surrounding the petiole inoculation site (Figure 6I).

Figure 6. Steps for root (A-H) and stem colonization (I) quantification - Place up to 100 mg of cut stem into a prepared grinding tube containing 4 metal beads and 900 μl of SMQ water.
- Grind the tissue using a Powerlyzer® bead-beater machine with the following settings:
Speed = 2,200 rpm, Time = 1 min 30 sec, Dwell = 4 min, Cycles = 2, Total time = 7 min - Serially dilute the homogenates 10-fold in 96-well plates by mixing 20 μl of each dilution with 180 μl of SMQ. For root samples: make five successive 1:10 dilutions per sample (100-10-4); for stem samples: make seven successive 1:10 dilutions per sample (100-10-6).
- Plate three 10-μl drops of each dilution onto CPG plates (with appropriate antibiotics). Dry the plates by opening the lid in a Biosafety Cabinet for approximately 15 min.
- Incubate the plates at 28 °C for 36-48h and count the colonies. Colonies should not be allowed to grow together.
Note: Normalize the final population size by the ratio of actual bacterial numbers in the original individual inoculum, as determined by dilution plating of the inoculum. Please see the attached Excel sheet for calculation.
- Xylem sap collection by root pressure
The following protocol is adapted from Goodger et al. (2005) and was used in Khokhani et al. (2017) and Lowe-Power et al. (2018).
For a basic experiment, we prepare 9 plants per treatment per time point (sampling is destructive). Sap will be combined into 3 pools, each containing sap from 3 plants. Occasionally sap cannot be collected from a plant due to uneven cutting, so it is prudent to prepare additional plants. Root pressure accumulates overnight when plants are in well-watered soil. Root pressure decreases over the course of the day. Therefore, sap collection should occur within several hours of light onset and plants must be well watered the evening before sap collection. Sap exudation rate depends on many variables, including soil moisture content, plant size, and disease state. In the experiments described in Lowe-Power et al. (2018), we harvested median volumes of 473 μl and 280 μl from healthy and wilt-symptomatic (disease index 1) plants that were 26-31 days old (Figure 7B).
Preparations:- One tube per plant to grind tissue to determine colonization (4 metal beads and 900 μl SMQ water)
- One tube per plant for the xylem sap collection (number, color code, etc.), pre-weighed, pre-cooled on ice. If possible, arrange all tubes and plants in the same parallel to avoid cross-contaminating samples.
- One tube per plant to sterilize the sap.
- One tube per pool.
- Set timer to 33 min.
- Detop the tomato plants with a sharp razor blade to yield a flat stump surface. If the stump surface is angled, sap will likely drip down the stem. For soil soak inoculated plants, cut ~1 cm above cotyledons. For cut-petiole inoculated plants, cut just above the inoculation site. Generally, it works well to detop plants in batches of 5 with 5 min intervals between batches.
- Allow the first droplet of sap to accumulate for ~2 ½ min. Then gently blot the stump dry with a tissue. Use a P200 pipette to wash the stump with sterile SMQ water, then blot dry again, to minimize cytosolic contamination.
- Collect xylem sap for 30 min, always using a fresh tip and pipetting the xylem sap of each plant into the corresponding tube, which is kept on ice. The sap should accumulate as a bubble due to surface tension (Figure 7A). If the stump looks wet, but sap is not accumulating, it is covertly dripping down the stem. Gently blotting the stump dry may restore surface tension to exuded sap and allow it to accumulate. We have included the estimation of xylem sap volume collected from the tomato plants (Figure 7B).
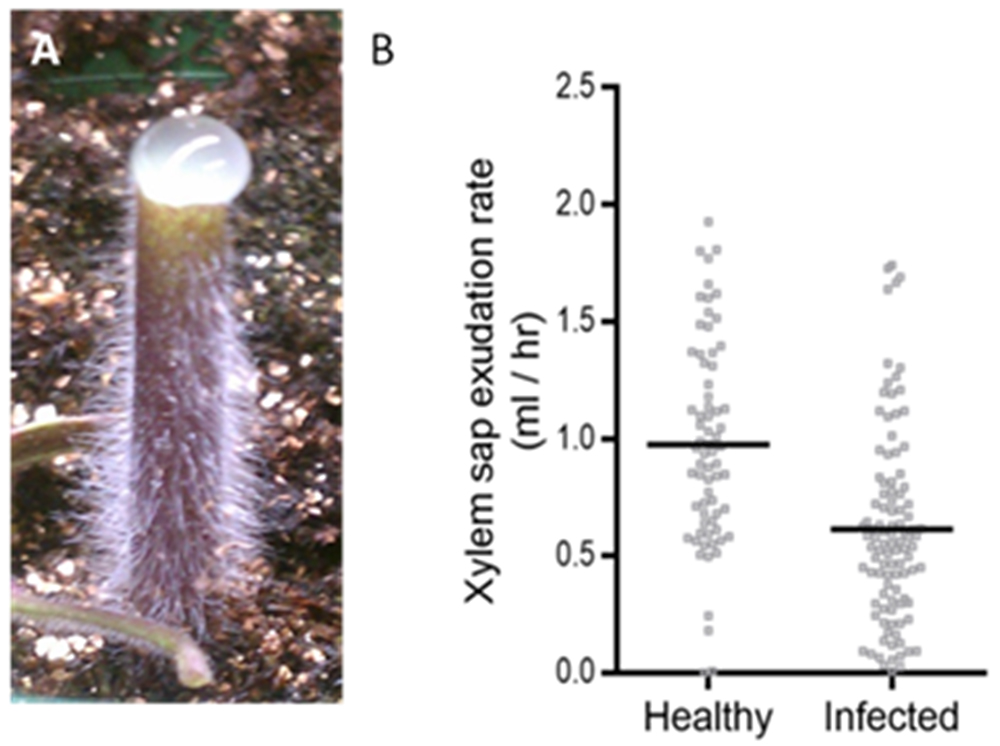
Figure 7. Collection of xylem sap. A. Xylem sap on detopped tomato plant. B. Sample dataset showing xylem sap volume from healthy and infected plants. Each dot represents the volume of sap collected from one plant. - Keep the top of the plants to take samples to quantify each plant’s bacterial colonization (collect a ~50-100 mg slice from the lowest part of the stem), as described above.
- Weigh each tube again to determine the xylem sap volume you gained from each plant (density is similar to water, so 1 mg = 1 μl).
- Pools should be composed of xylem sap from plants producing high, medium and low amount of xylem sap. For pooling, combine equal volumes of xylem sap of each plant (e.g., 100 μl). Filter-sterilize the xylem sap from each plant into a fresh 1.5 ml tube using a 1 ml syringe and a 4 mm diameter 0.22 μm filter (4 mm size filters reduce sample loss).
- This xylem sap can be used as medium for growth curve assays or for metabolite analysis. Bacterial growth assays can be performed with 50 μl of sap in Corning 96-well half-area microplates. The amount of sap needed for metabolite analysis should be empirically determined based on the concentration of target metabolites and sensitivity of the detection method.
- Root attachment to tomato seedlings
The following protocol has been used in Tran et al. (2016b) and Khokhani et al. (2017).- Surface sterilize the tomato seeds by soaking them in 2% bleach for 5 min, followed by 70% ethanol for 5 min, then six rinses with SMQ water.
- With the help of sterile forceps in a biosafety cabinet, transfer the sterilized seedlings by spreading them out on to 1% agar Petri plates overlaid with a sterile WhatmanTM filter paper.
- Seal the Petri plates with MicroporeTM tape and cover with aluminum foil to exclude light.
- Incubate the plates at 28 °C for 4 days. In the meantime, prepare 1% agar square plates, CPG + TZC + required antibiotic, bead-beater tubes with 4 metal beads and 300 μl of SMQ water.
- On the 4th day, mark the back of the square plates with sharpie as shown here (Figure 8).
Figure 8. Square plates marked with 2 cm area in which the roots of tomato seedlings are arranged - In the biosafety cabinet, arrange the tomato seedlings so that their roots span the 2-cm length (from the root tip). Approximately 40 tomato seedlings can be accommodated in one square plate, with 20 on each side.
- Prepare the bacterial inoculum as described in Steps D2-D6.
- Dilution plate the inoculum and incubate the plates at 28 °C to confirm inoculum density.
- Next, use a micropipette to carefully distribute 10 μl of the OD600 = 0.001 bacterial suspension (~10,000 cells) over the 2-cm length of each root in the square plate.
- Incubate the square plates at room temperature for 2 h.
- After two hours, use a sterile scalpel to cut out the 2-cm length indicated by the sharpie marks for all 40 roots.
- Transfer all the cut roots to a Petri dish containing SMQ water and swirl the plate to remove loosely attached bacterial cells. After the first rinse, transfer all the roots to another Petri plate containing SMQ water and rinse again.
- Next, transfer the roots on to a new paper towel and gently pat dry the roots. Transfer 4 roots at one time.
- After pat-drying, transfer each group of 4 roots into a prepared bead-beater tube containing metal beads and 300 μl SMQ water. For 40 roots, you will have 10 technical replicates per biological replicate per treatment.
- Place the bead-beater tubes containing cut roots along with metal beads in the Powerlyzer® using the following settings: Speed = 2,200 rpm, Time = 1 min 30 sec, Dwell = 4 min, Cycles = 2, Total time = 7 min.
- After grinding, plate the homogenates along with their 1:10, 1:100, 1:1,000 serial dilutions for each sample.
- After 48 h, count the colonies to determine the number of bacterial cells in the original inoculum and the number of bacteria attached to the roots.
- Calculate the percent attachment by dividing the number of bacterial cells recovered from root by the number of cells present in the inoculum, per cm of roots.
Note: Please see the attached Excel sheet for calculation.
Data analysis
Figure 9 represents a disease progress assay from Dalsing et al. (2015). Plants were rated daily for 14 days using the 0 to 4 disease index as described above. Data presented are mean results from 3 to 4 independent assays, each containing 10 plants per strain. Error bars indicate standard errors of the means. Disease progress curves of the ΔaniA and ΔhmpX mutants were significantly different from those of the WT strain (P < 0.001, repeated measures ANOVA, PRISM Graphpad software).
The protocols presented here generate quantitative data that demand thoughtful, nuanced analysis. Fortunately, a recent paper (Schandry, 2017) offers a detailed practical guide to analyzing data on R. solanacearum infection, including the relevant R code.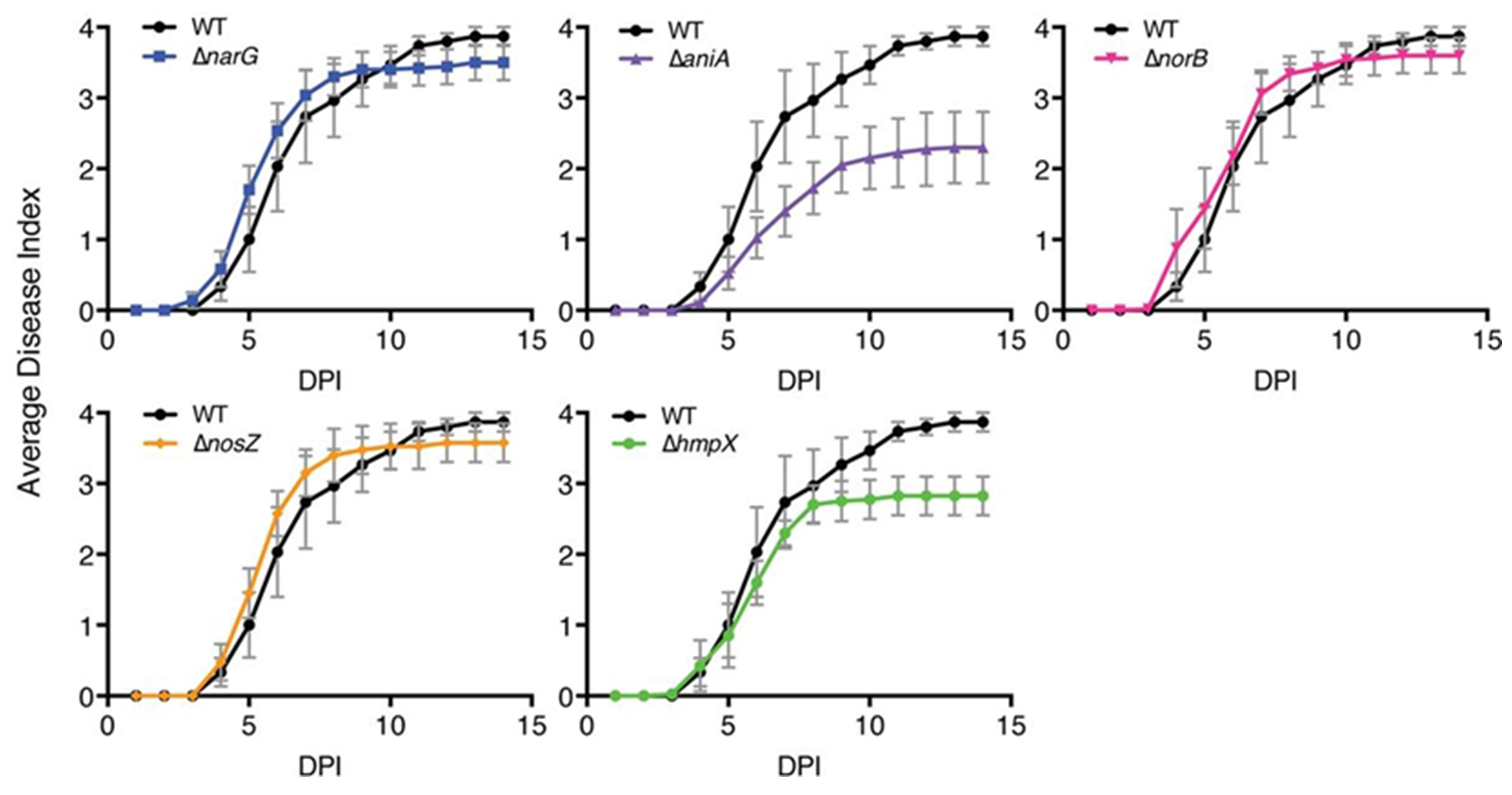
Figure 9. Sample disease progress graphs from one of the virulence assays in Dalsing et al. (2015)
Notes
- Calibrate your pipettes regularly, especially the P10 used for dilution plating.
- With every new experiment, use a new bottle of SMQ water to avoid any contamination.
- Check your strain stocks regularly for spontaneous phcA mutants.
- Do not inoculate using colonies from CPG plates that have grown for more than 4 days after streaking out from water or freezer stocks. “Old” colonies have often lost virulence.
Recipes
- Casamino acid-peptone-glucose (CPG) agar (1 L)
5 g Glucose
10 g Peptone
1 g Casamino acids
1 g Yeast extract
Adjust to pH 6.5-7.0 with 1 M KOH
16 g Agar
Autoclave at 121 °C, 20 min
1 ml of 1% 2,3,5-triphenyl tetrazolium chloride (TZC) (dissolved in water, filter-sterilized and added into CPG before pouring the plates) - CPG broth (1 L)
5 g Glucose
10 g Peptone
1 g Casamino acids
1 g Yeast extract
Adjust to pH 6.5-7.0 with KOH
Autoclave - Modified Hoagland's solution
Mixture
1.1 M KNO3
0.2 M KH2PO4
0.92 M MgSO4
For watering, use 4 ml/L water
Calcium Nitrate
1.29 M Ca(NO3)2•4H2O
For watering, use 5 ml/L water
Micronutrients
0.4626 M H3BO3 (Boric acid)
0.0915 M MnCl2•4H2O (Manganese Chloride)
0.0077 M ZnSO4•7H2O (Zinc Sulfate)
0.0032 M CuSO4•5H2O (Copper Sulfate)
0.0007 M (NH4)6Mo7O24 (Molybdic acid)
For watering, use 4 ml of the stock/L water
Iron
FeSO4•7H2O (Ferrous Sulfate) 25.02 g/18 L of water
Na2EDTA (Ethylenediaminetetraacetic acid) 33.48 g/18 L
Dissolve the above in 4 L of deionized RO water. Heat to 80 °C for one hour. Let cool to room temperature and add deionized RO water to 18 L
For watering, use 4 ml/L water
Acknowledgments
We thank Patrizia Ricca for capturing the images of soil soak, root and stem colonization. We also thank all the authors of whose papers have been cited for adapting and/or modifying their protocols to perform the respective experiments.
Competing interests
The authors declare no conflict of interest.
References
- Alvarez, B., Lopez, M. M. and Biosca, E. G. (2008). Survival strategies and pathogenicity of Ralstonia solanacearum phylotype II subjected to prolonged starvation in environmental water microcosms. Microbiology 154(Pt 11): 3590-3598.
- Brown, D. G. and Allen, C. (2004). Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol Microbiol 53(6): 1641-1660.
- Colburn-Clifford, J. M., Scherf, J. M. and Allen, C. (2010). Ralstonia solanacearum Dps contributes to oxidative stress tolerance and to colonization of and virulence on tomato plants. Appl Environ Microbiol 76(22): 7392-7399.
- Dalsing, B. L. and Allen, C. (2014). Nitrate assimilation contributes to Ralstonia solanacearum root attachment, stem colonization, and virulence. J Bacteriol 196(5): 949-960.
- Dalsing, B. L., Truchon, A. N., Gonzalez-Orta, E. T., Milling, A. S. and Allen, C. (2015). Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment. MBio 6(2): e02471.
- Denny, T. (2007). Plant pathogenic Ralstonia species. In: Gnanamanickam, S. S. (Ed.). Plant-Associated Bacteria. Springer 573-644.
- Elphinstone, J. G. (2005). The current bacterial wilt situation: a global overview. In: Allen, C., Prior, P. and Hayward, A. C. (Eds.). Bacterial Wilt Disease and the Ralstonia solanacearun Species Complex. APS Press 9-28.
- Genin, S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol 187(4): 920-928.
- Genin, S. and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50: 67-89.
- Goodger, J. Q. D., Sharp, R. E., Marsh, E. L. and Schachtman, D. P. (2005) Relationships between xylem sap constituents and leaf conductance of well‐watered and water‐stressed maize across three xylem sap sampling techniques. J Exp Bot 56(419): 2389-2400.
- Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29: 65-87.
- Huang, Q. and Allen, C. (2000). Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol Mol PlantPathol 57(2):77-83.
- Jacobs, J. M., Babujee, L., Meng, F., Milling, A. and Allen, C. (2012). The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. MBio 3(4): e00114-12.
- Khokhani, D., Lowe-Power, T. M., Tran, T. M. and Allen, C. (2017). A single regulator mediates strategic switching between attachment/spread and growth/virulence in the plant pathogen Ralstonia solanacearum. MBio 8(5): e00895-17.
- Lowe, T. M., Ailloud, F. and Allen, C. (2015). Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Mol Plant Microbe Interact 28(3): 286-297.
- Lowe-Power, T. M., Hendrich, C. G., von Roepenack-Lahaye, E., Li, B., Wu, D., Mitra, R., Dalsing, B. L., Ricca, P., Naidoo, J., Cook, D., Jancewicz, A., Masson, P., Thomma, B., Lahaye, T., Michael, A. J. and Allen, C. (2018). Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ Microbiol 20(4): 1330-1349.
- Lowe-Power, T. M., Jacobs, J. M., Ailloud, F., Fochs, B., Prior, P. and Allen, C. (2016). Degradation of the plant defense signal salicylic acid protects Ralstonia solanacearum from toxicity and enhances virulence on tobacco. MBio 7(3): e00656-16.
- Macho, A. P., Guidot, A., Barberis, P., Beuzón and Genin, S. (2010). A competitive index assay identifies several ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. MPMI 23(9):1197-1205.
- Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., Dow, M., Verdier, V., Beer, S. V., Machado, M. A., Toth, I., Salmond, G. and Foster, G. D. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13(6): 614-629.
- Peyraud, R., Cottret, L., Marmiesse, L., Gouzy, J. and Genin, S. (2016). A Resource Allocation Trade-off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathog 12(10): e1005939.
- Saile, E., McGarvey, J. A., Schell, M. A. and Denny, T. P. (1997). Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87(12): 1264-1271.
- Schandry, N. (2017). A practical guide to visualization and statistical analysis of R. solanacearum infection data using R. Front Plant Sci 8: 623.
- Tans-Kersten, J., Huang, H. and Allen, C. (2001). Ralstonia solanacearum needs motility for invasive virulence on tomato. J Bacteriol 183(12): 3597-3605.
- Tran, T. M., MacIntyre, A., Hawes, M. and Allen, C. (2016b). Escaping underground nets: Extracellular DNases degrade plant extracellular traps and contribute to virulence of the plant pathogenic bacterium Ralstonia solanacearum. PLoS Pathog 12(6): e1005686.
- Tran, T. M., MacIntyre, A., Khokhani, D., Hawes, M. and Allen, C. (2016a). Extracellular DNases of Ralstonia solanacearum modulate biofilms and facilitate bacterial wilt virulence. Environ Microbiol 18(11): 4103-4117.
- Weibel, J., Tran, T. M., Bocsanczy, A. M., Daughtrey, M., Norman, D. J., Mejia, L. and Allen, C. (2016). A Ralstonia solancearum strain from Guatemala infects diverse flower crops, including new asymptomatic hosts Vinca and Sutera, and causes symptoms in geranium, mandevilla vine, and new host Afircan daisy (Osteospermum ecklonis). Plant Health Prog 17: 114-121.
- Wicker, E., Grassart, L., Coranson-Beaudu, R., Mian, D., Guilbaud, C., Fegan, M. and Prior, P. (2007). Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl Environ Microbiol 73(21): 6790-6801.
- Yao, J. and Allen, C. (2006). Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol 188(10): 3697-3708.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Khokhani, D., Tuan, T. M., Lowe-Power, T. M. and Allen, C. (2018). Plant Assays for Quantifying Ralstonia solanacearum Virulence . Bio-protocol 8(18): e3028. DOI: 10.21769/BioProtoc.3028.
Category
Microbiology > Microbe-host interactions > Bacterium
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










