- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
6-hydroxydopamine (6-OHDA) Oxidative Stress Assay for Observing Dopaminergic Neuron Loss in Caenorhabditis elegans
Published: Vol 8, Iss 18, Sep 20, 2018 DOI: 10.21769/BioProtoc.3025 Views: 7515
Reviewed by: Khyati Hitesh ShahAnand Ramesh PatwardhanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Simultaneous Monitoring Cytoplasmic Calcium Ion and Cell Surface Phosphatidylserine in the Necrotic Touch Neurons of Caenorhabditis elegans
Yoshitaka Furuta and Zheng Zhou
Oct 20, 2021 2623 Views

Live-cell Imaging and Analysis of Germline Stem Cell Mitosis in Caenorhabditis elegans
Réda M. Zellag [...] Abigail R. Gerhold
Jan 5, 2022 4694 Views

SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
Elise van der Salm [...] Suzan Ruijtenberg
Oct 20, 2025 2280 Views
Abstract
The nematode Caenorhabditis elegans is a powerful genetic model that can be used to investigate neuronal death. Research using C. elegans has been crucial to characterize cell death programmes that are conserved in mammals. Many neuronal signaling components, such as those mediating dopaminergic neurotransmission, are preserved as well. Dopaminergic neurons are progressively lost in Parkinson’s disease and an important risk factor to develop this disease appears to be oxidative stress, the increased occurrence of highly reactive oxygen species. Oxidative stress-induced dopaminergic neurodegeneration is mimicked in animal models by treatment with 6-hydroxydopamine (6-OHDA), a dopamine analog, which is specifically taken up into dopaminergic neurons. After exposing C. elegans to 6-OHDA, the loss of fluorescently labeled dopaminergic neurons can be easily monitored. An organisms’ sensitivity to oxidative stress is thought to be influenced by basal levels of intrinsic oxidative stress and the ability to counteract oxidative stress and oxidative stress-induced damage. The C. elegans ‘6-OHDA model’ led to the discovery of novel genes that are required to protect dopaminergic neurons and it has helped to determine the effects of conserved cell death and cell engulfment pathways in dopaminergic neurodegeneration. Here, we describe a simple protocol that allows for the easy detection of dopaminergic neuron loss after 6-OHDA treatment in C. elegans.
Keywords: C. elegansBackground
The gradual loss of dopaminergic neurons can be recapitulated in animal models following exposure to the oxidative stress-inducing drug 6-hydroxydopamine (6-OHDA) (for review Schober, 2004). In contrast to other neurodegenerative drugs such as MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine), 6-OHDA is safer to handle as it does not pass the blood-brain-barrier. 6-OHDA is a hydroxylated dopamine analog, which is specifically taken up into dopaminergic neurons by the dopamine transporter and blocks complex I of the respiratory chain (Schober, 2004). The resulting formation of reactive oxygen species is thought to trigger 6-OHDA-induced neurodegeneration (Schober, 2004).
6-OHDA exposure of the nematode Caenorhabditis elegans leads to the selective loss of dopaminergic neurons (Nass et al., 2002) and components of dopaminergic neurotransmission are highly conserved compared to mammals (for review Nass et al., 2001). C. elegans hermaphrodites possess eight dopaminergic neurons: four CEP (cephalic sensilla) and two ADE (anterior deirids) dopaminergic neurons in the head, and two PDE (posterior deirids) dopaminergic neurons in the midbody (Sulston et al., 1975). These neurons can be specifically labeled by expression of a fluorescent protein driven by the promoter of the dat-1 dopamine transporter (Nass et al., 2002). The C. elegans 6-OHDA model can be used to understand how dopaminergic neurons maintain their integrity when subjected to oxidative stress.
The first published 6-OHDA intoxication protocol for C. elegans used high concentrations of 6-OHDA elicit dopaminergic neurodegeneration in wild-type animals (Nass et al., 2002). Mutation of the dopamine transporter dat-1, which is required for neuronal 6-OHDA uptake, was shown to confer 6-OHDA resistance (Nass et al., 2002). After 6-OHDA exposure during larval stages (L3 and L4), dopaminergic neurodegeneration was scored in a low-throughput manner by mounting adult animals on cover slide (Nass et al., 2002; Tucci et al., 2011). We adapted the protocol to screen for mutants that are hypersensitive to 6-OHDA exposure by using lower 6-OHDA concentrations that do not elicit neurodegeneration in wild-type animals. We expose synchronized L1 stage larvae in 96-well plates and score adults directly on agar plates, allowing for high-throughput screening. This approach led to the characterization of the tetraspanin gene tsp-17, the neuroligin-like gene glit-1 and the transthyretin-related gene ttr-33, all of which protect C. elegans dopaminergic neurons from 6-OHDA-induced neurodegeneration (Masoudi et al., 2014; Offenburger et al., 2018a and 2018b). The C. elegans 6-OHDA assay was further used to describe the roles of known stress response and cell death pathways in oxidative stress-induced dopaminergic neurodegeneration (Nass et al., 2002; Tóth et al., 2007; Offenburger et al., 2018a and 2018b).
The protocol can also be used for acute liquid exposure to other soluble compounds such as the oxidative stress-inducing drug paraquat (Offenburger et al., 2018a and 2018b). The procedures we describe here are generally useful to test if compounds influence dopaminergic neuron death.
Materials and Reagents
- Latex gloves
- Lab coat
- Platinum wire (e.g., CVS10 replacement platinum wire, 50 cm length x 0.2 mm diameter, Sigma-Aldrich, catalog number: EP1330 )
- High recovery filter pipette tips (e.g., Corning, Axygen® Maxymum Recovery®, catalog number: T-200-C-L-R-S )
- Bench surface protector sheets (GE Healthcare, Whatman®, catalog number: 2300-916 )
- Disposable spatulas (e.g., Disposable smartSpatula, LevGo, catalog number: 17251 )
- 1.5 ml screw cap tubes (sterile, graduated, conical, e.g., STARLAB, catalog number: E1415-2231 )
- 96-well plates (Tissue culture plates, round bottom, clear, sterile, TPP Techno Plastic Products, catalog number: 92097 )
- Only if filtering of L1 larval stages required: Nylon net filter, hydrophilic, 5 μm pore size (e.g., Merck, catalog number: NY0509050 with 90 mm diameter, or catalog number: NY0502500 with 25 mm diameter)
- 10 cm plastic Petri dishes
- Wet paper towel
- E. coli OP50 [Caenorhabditis Genetics Centre (CGC), University of Minnesota, Dept of GCD, 6-160 Jackson Hall, 321 Church Street S.E. Minneapolis, MN 55455, https://cgc.umn.edu/]
- Experimental control strains available at the C. elegans Genetics Centre (CGC), University of Minnesota, https://cgc.umn.edu/):
- TG2435 wild type-backcrossed BY200 derivate, vtIs1[pdat-1::gfp; rol-6] V (while pdat-1::gfp is always expressed, the penetrance of the roller phenotype is very low, only rarely detectable)
- TG2400 dat-1(ok157) III; vtIs1V
- TG4100 vtIs1 V; glit-1(gt1981) X
- TG2436 vtIs1 V; tsp-17(tm4995) X
- G4103 ttr-33(gt1983) V; vtIs1 V
- M9 buffer (He, 2011)
- Nematode growth medium (NGM) agar (He, 2011)
- LB (Luria-Bertani) liquid medium (see Cold Spring Harbor Protocols, 2006)
- 6-hydroxydopamine (6-OHDA) hydrochloride (Sigma-Aldrich, catalog number: H4381 ) (keep at -20 °C in the dark, prepare stock solution freshly)
- L-ascorbic acid (Sigma-Aldrich, catalog number: A5960 , ≥ 90%) (store at room temperature in the dark, wrap aliquots with aluminum foil)
- 200 mM ascorbic acid solution (freshly prepared, see Recipes)
- 10 mM 6-OHDA solution (freshly prepared, see Recipes)
Equipment
- Pipettes
- Metal inoculation loop
- Precision scales
- Temperature-controlled shaker (e.g., Eppendorf, model: ThermoMixer® R )
- Temperature-controlled incubator (20 °C)
- Fume hood
- Vortex Mixer (e.g., Scientific Industries, model: Vortex-Genie 2 )
- Ethanol burner (e.g., DWK Life Sciences, WheatonTM Alcohol Burner, catalog number: 237070 )
- Stereomicroscope with fluorescence (e.g., Leica)
Software
- R studio (version 1.0.44)
Procedure
- Preparation
- Prepare NGM plates and dry overnight at 37 °C. Store unseeded plates at 4 °C.
- Transfer the plates to room temperature before covering them with ca. 1 ml of OP50 E. coli. We prepare E. coli cultures by inoculating a single colony of OP50 in 500 ml LB medium and overnight incubation at 37 °C.
- Dry these seeded plates overnight and store them at 20 °C.
- Maintain C. elegans at standard conditions at 20 °C (Brenner, 1974) on 10 cm plastic Petri dishes containing nematode growth medium (NGM) agar and OP50 bacteria food.
Note: For the 6-OHDA experiments, the N2-derived BY200 C. elegans strain can be used as a wild-type control, dat-1 as a 6-OHDA-resistant strain (Nass et al., 2002) and the mutant strains tsp-17 (Masoudi et al., 2014), glit-1 (Offenburger et al., 2018b) or ttr-33 (Offenburger et al., 2018a) as 6-OHDA-sensitive mutants.
- 6-OHDA assay (Figure 1)

Figure 1. Overview of daily schedule of 6-OHDA assay. Please refer to text for details.- One day before the 6-OHDA assay, pick gravid C. elegans (in adult stage and full of eggs) into a 96-well plate containing 70 μl M9 buffer per well. Only 30 μl of these 70 μl will be used for the 6-OHDA treatment, but as a substantial amount of the liquid will evaporate, it is necessary to prepare a larger volume. It is best practice to select healthy day 1 or day 2 adults for all conditions and replicates.
Note: We recommend avoiding wells on the edge of the plate (rows A and H, and columns 1 and 12, respectively), as liquids in the outermost wells show increased evaporation. - Transfer 10 adults into each well using a platinum wire, minimizing as little bacteria carryover as possible, as bacterial growth in the 96-well plates would compromise the synchronization of C. elegans L1 stage larvae.
Note: If necessary, first transfer worms to an empty NGM plate for a few seconds and then move clean animals that have crawled away from the bacteria to the 96-well plate. - Prepare a technical duplicate for each experiment (i.e., two wells per condition) and perform at least two biological replicates on different days. We encourage randomizing the sequence of the analyzed strains (Figure 2).
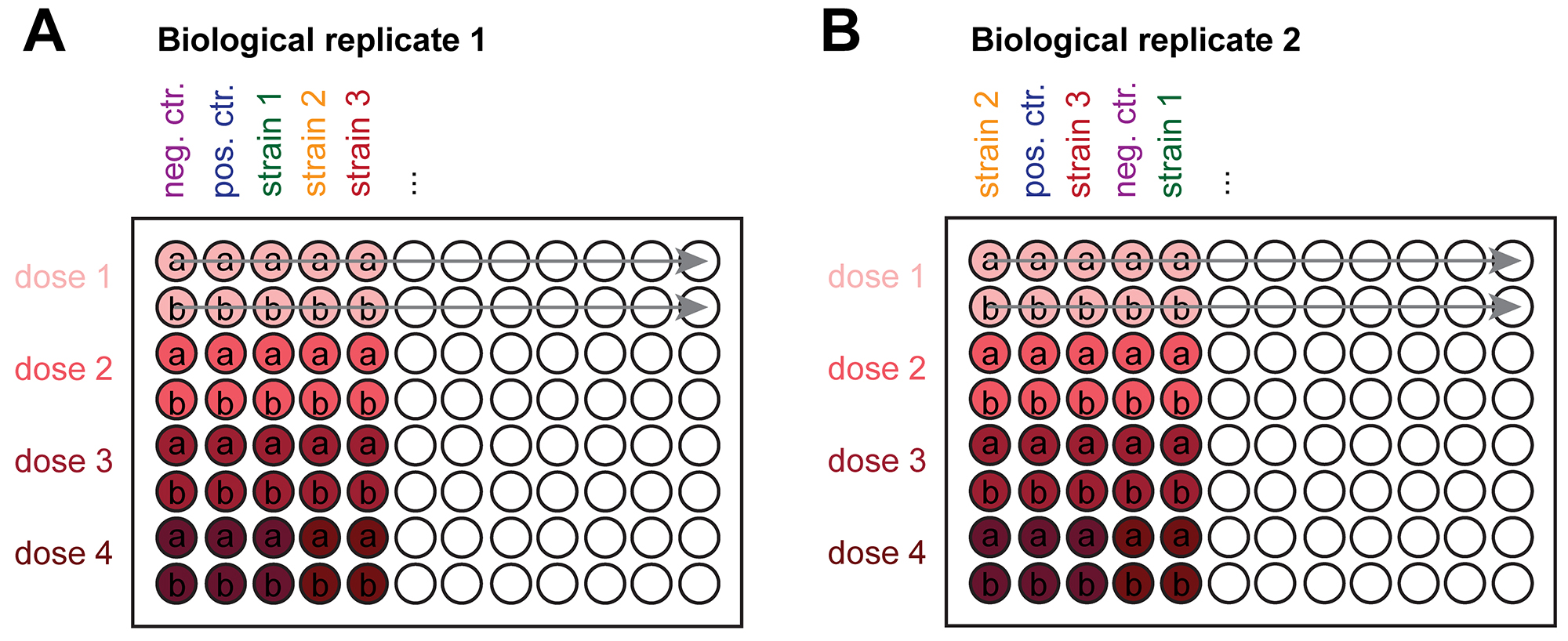
Figure 2. Schematic representation of oxidative stress assay design. C. elegans are exposed in liquid in 96-well plates. The assay is run in technical duplicate (rows a + b per dose) including negative and positive controls. We encourage randomizing the position of control strains between the biological replicates (A + B). The biological replicates must be performed on separate days. The red color gradient indicates the increasing dose of the drug. We commonly used doses of 0.75 mM, 10 mM, 25 mM and 50 mM 6-OHDA. We recommend avoiding wells on the edge of the plate (row A and H and column 1 and 12), as liquids in the outermost wells show increased evaporation. We maintain the same pipetting sequence (indicated with a grey arrow) when adding compounds to ensure equal exposure of all tested strains. - Incubate the 96-well plates in a temperature-controlled shaker at 500 rpm at 20 °C for 24-40 h. As food is absent in the wells, the eggs laid by these adults will hatch yet remain in the L1 larval stage, thus providing a developmentally synchronous culture. We use 30 h of incubation time for all our experiments and encourage standardizing this time interval as starvation renders the animals more resistant to 6-OHDA treatment (González-Hunt et al., 2014; Offenburger et al., 2018b). After 30 h, several hundred L1s will accumulate in the wells, of which only around 200 will be used in the assay and only 50-100 randomly selected animals will be scored after treatment.
- On the day of 6-OHDA treatment, first prepare the recovery plates. Label NGM agar-containing 6 cm plates in the back with the respective letter/number code from the 96-well plate and streak a line of OP50 E. coli on the NGM using a metal inoculation loop (Figure 3A).
Notes:- Other items can be used to streak the line of OP50. However, it is important not to damage the agar surface as otherwise animals might bury into the agar and will thus be lost for scoring.
- To ensure the food does not run out when analyzing a large number of animals, it is possible to streak concentrated bacteria (OP50 E. coli centrifuged for 5 min at > 15,000 x g and resuspended in 1/10 volume of LB medium).
- Immediately before treating the animals, prepare a 200 mM ascorbic acid stock solution and a 5x stock solution of 6-OHDA in MilliQ water in screw cap tubes. 10 μl ascorbic acid stock solution and 10 μl 6-OHDA stock solution are required per well in a total volume of 50 μl. The 1:5 dilution results in a final concentration of 40 mM of ascorbic acid and 1x 6-OHDA (we commonly used, 0.75, 10, 25 or 50 mM 6-OHDA in our experiments). Ascorbic acid is an antioxidant, which prevents auto-oxidation of 6-OHDA.
Note: When handling 6-OHDA, wear double gloves and a lab coat, use disposable filter tips and work under the fume hood. Use a disposable bench cover in the fume hood and put a wet paper towel in the area in which 6-OHDA is handled such that spilled 6-OHDA is oxidized immediately. - Weigh ascorbic acid and 6-OHDA in the screw cap tubes using disposable spatulas and precision scales in the fume hood. To minimize handling time, weigh an approximate, slightly higher amount of compound and adapt the total volume accordingly.
- Separate 6-OHDA waste materials and dispose of properly. Try to work with the same batch of 6-OHDA as efficacies can vary between production lots. Vortex both the ascorbic acid solution and the 6-OHDA solution thoroughly for approximately 1 min until flakes are dissolved and the solutions appear homogenous.
Note: The screw caps help to ensure that that the tubes stay closed during mixing. If working with several different concentrations or with small concentrations, prepare dilution series from the same stock solution. - Aliquot 30 μl of worms in M9 buffer (corresponding to over 200 animals) into new wells using non-adhesive filter tips to minimize animal loss.
Note: A precise multi-channel pipette can be used to transfer animals. Technical replicates are not intermixed at this stage. If animal density varies greatly between the wells, it can be adjusted by diluting the worms in M9 buffer to produce a total volume of 30 μl. - Check with a stereomicroscope if a sufficiently high number of animals (at least 50 animals per well) have been transferred before adding the compounds.
- Then add 10 μl ascorbic acid solution and knock 96-well plate on the bench or centrifuge the plate shortly to make sure the contents are mixed properly before adding 6-OHDA.
Note: No drops of ascorbic acid solution should remain on the wall of the well. Shake the plate shortly to ensure equal distribution of the ascorbic acid in the wells. To prevent auto-oxidation of 6-OHDA, the ascorbic acid must be added first. A precise multi-channel pipette can be used to dispense reagents. - Add 10 μl 5x 6-OHDA solution and again ensure the solutions are properly mixed. Shake the multiwell plates for 1 h at 500 rpm at 20 °C.
- Stop the incubation by adding 150 μl M9 buffer. This buffer will oxidize the 6-OHDA and the solution will turn pink to dark red. The intensity of the color after addition of the buffer is a good indicator of the 6-OHDA concentration used.
- Pipet the total of 200 μl of worms into oxidized 6-OHDA solution on the NGM plates opposite the bacterial stripe, directly on top of the plate label (Figure 3B).
- Open the lids and let the plates dry under the fume hood. When the 6-OHDA solution has soaked into the NGM, remove the adult animals and eggs with a platinum wire such that only animals that were exposed at L1 stage remain on the plate (Figure 3C). Unless working with mutant strains that exhibit a developmental defect, there are usually no eggs or very few eggs present as all of progeny will have hatched in the 24-40 h incubation time before the assay. The L1 larvae remaining on the scoring plates will migrate towards the bacterial streak, which provides the food source (Figure 3D).
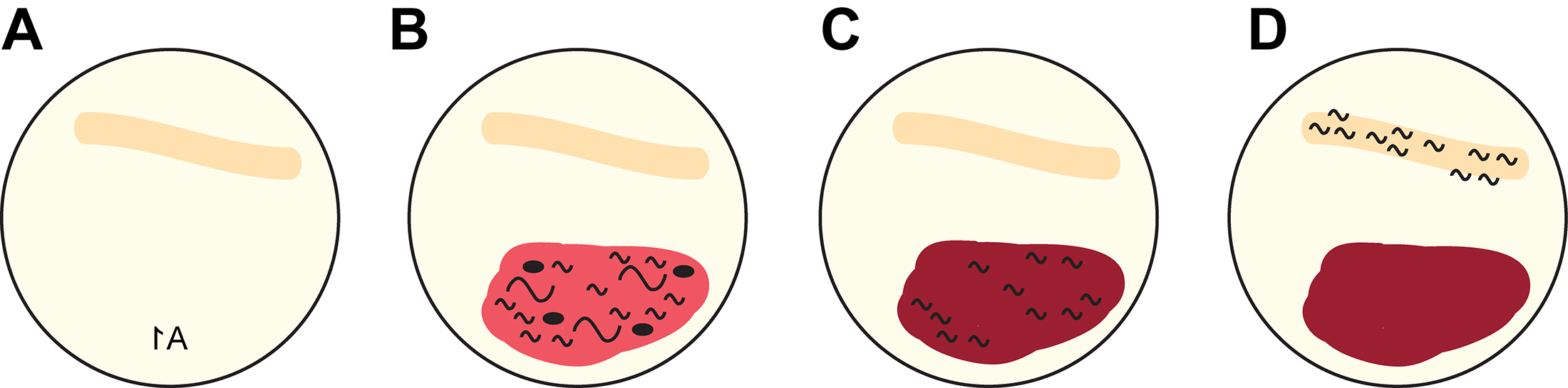
Figure 3. Layout of recovery plates. A. 6 cm NGM Petri dish labeled in the back with the well number (e.g., 1A) and inoculated with a stripe of OP50 E. coli (orange streak). B. The mixture of adult and L1 stage animals and unhatched eggs in oxidized 6-OHDA (red liquid) is added to the plate on the side opposite to the bacterial streak. C. Adult animals and unhatched eggs are removed with a platinum wire. D. The remaining L1 stage larvae will migrate towards the bacterial streak, allowing for convenient scoring. - Check the plates daily to make sure the bacterial food does not run out. In case there is little food left, add a drop (ca. 50-100 μl) of concentrated E. coli to prevent animals from starvation. Dopaminergic neurodegeneration can be observed 24, 48 or 72 h after exposure. The experimenter should be blinded to the genotype. C. elegans dopaminergic head neurons (Figure 4A) show differential sensitivity to 6-OHDA, with CEP neurons being more sensitive than ADE neurons (Nass et al., 2002; Tucci et al., 2011). PDE dopaminergic neurons in the midbody only appear after the L1 larval stage and are not scored in our experiments. Score at least 50 animals per replicate plate, resulting in a total number of at least 100 scored animals for each condition. Animals were selected randomly for scoring. We use four categories for scoring: (1) CEP + ADE neurons intact, (2) CEP neurons partially compromised and ADE neurons intact, (3) only ADE neurons remaining, and (4) no CEP or ADE neurons left (legend in Figure 4B, Table 1). This assay can be used for genetic screens to look for hypersensitive mutants (Figure 4B), to analyze epistasis between different mutations at differing concentrations of 6-OHDA (Figure 4C), or to analyze animals upon exposure to other soluble compounds such as paraquat (Figure 4D).
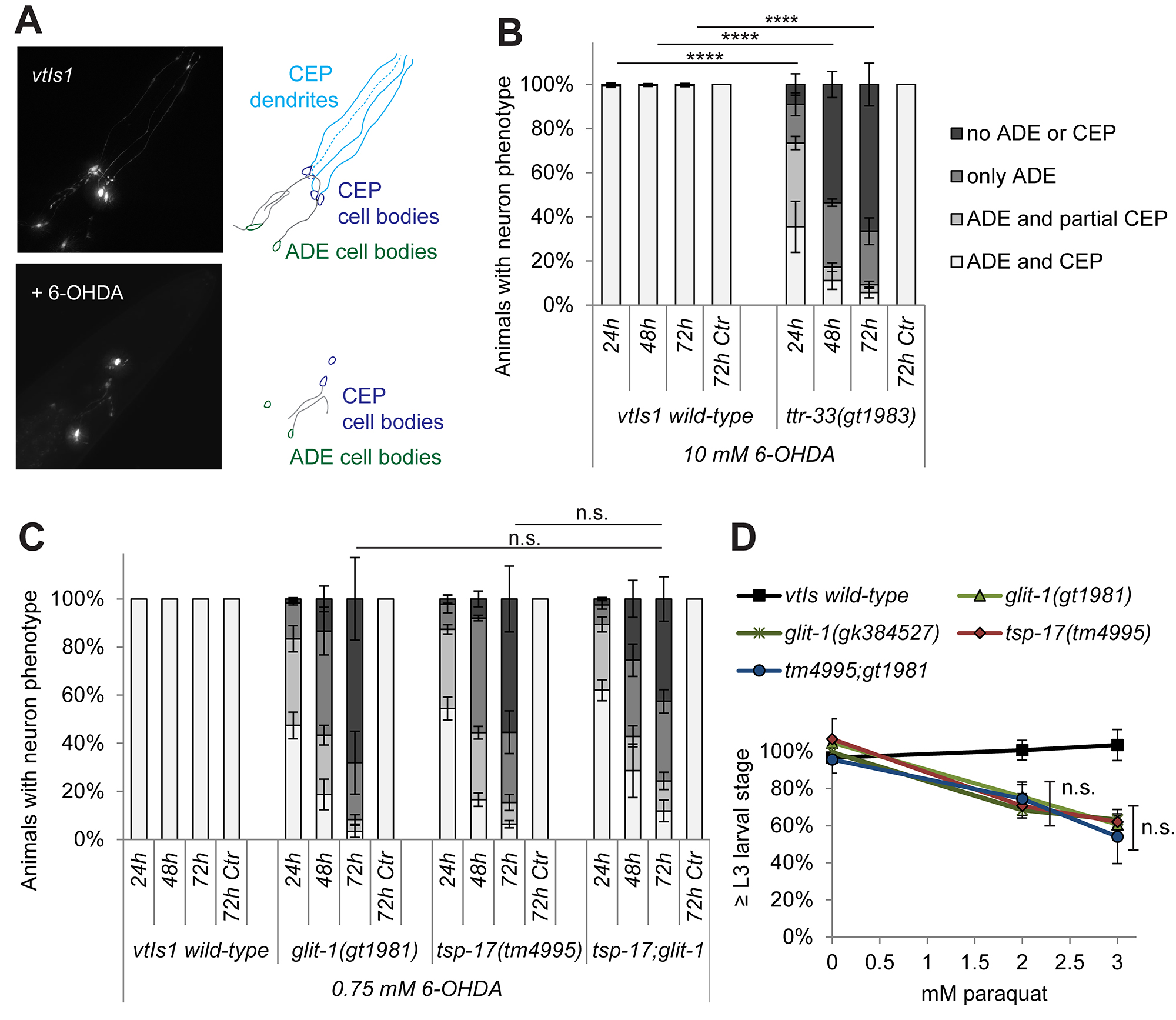
Figure 4. Representative readouts from oxidative stress assay. Panels A and B are adapted from Offenburger et al. (2018a) and panel C and D are adapted from Offenburger et al. (2018b). A. Fluorescently labeled C. elegans dopaminergic head neurons in the wild-type strain carrying the vtIs1 transgene (left) and schematic labeling (right) without (top) and with exposure to 50 mM 6-OHDA (‘+6-OHDA’, bottom) 72 h after treatment. ADE neurons are located posterior to CEP neurons. B. Remaining dopaminergic head neurons 24, 48 and 72 h after exposure to 10 mM 6-OHDA and 72 h after control treatment with ascorbic acid only (‘72h Ctr’) in BY200 wild-type animals and in the isolated hypersensitive mutant ttr-33(gt1983). Error bars = SEM of 3 biological replicates, each with 60-120 animals per strain. Total number of animals per condition n = 30 for the ‘72h Ctr’ and n = 220-340 for all the other conditions (**** P < 0.0001; G-Test comparing wild-type and mutant data of the same time point). C. Dopaminergic head neurons in wild-type and glit-1 and tsp-17 single and double mutants 24, 48 and 72 h after treatment with 0.75 mM 6-OHDA and 72 h after control treatment with ascorbic acid only (‘72h Ctr’). Error bars = SEM of 2-3 biological replicates, each with 60-115 animals per strain. Total number of animals per condition n = 60 for the ‘72h Ctr’ condition and n = 180-350 for all the other conditions (n.s. P > 0.05; G-Test). D. Percentage of animals that developed to L3 stage 24 h after 1 h incubation with 50 mM paraquat. Error bars = SEM of 3 biological replicates, each with 85-380 animals per strain and concentration. Total number of animals per condition n = 340-830 (n.s. P > 0.05; two-tailed t-test comparing the tsp-17(tm4995) and glit-1(gt1981) single mutants to the tsp-17(tm4995);glit-1(gt1981) double mutant at 2 mM and 3 mM paraquat).
Table 1. Example scoring for strains a and b. Number of animals in each category (sum of technical duplicate).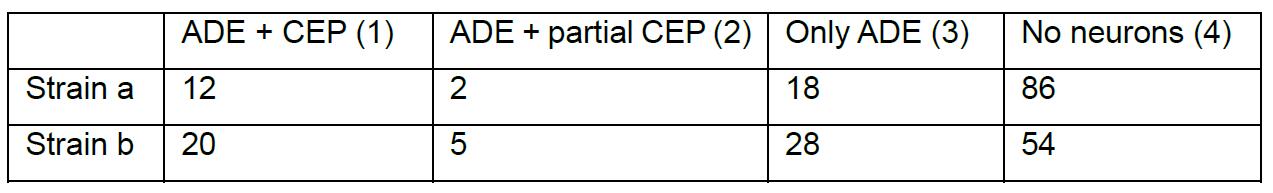
- One day before the 6-OHDA assay, pick gravid C. elegans (in adult stage and full of eggs) into a 96-well plate containing 70 μl M9 buffer per well. Only 30 μl of these 70 μl will be used for the 6-OHDA treatment, but as a substantial amount of the liquid will evaporate, it is necessary to prepare a larger volume. It is best practice to select healthy day 1 or day 2 adults for all conditions and replicates.
Data analysis
We visualized scoring results with the ‘100% Stacked Colum’ chart in Microsoft Excel. We added up numbers scored in technical duplicates and excluded data points for which the standard deviation between the technical duplicates was higher than 20% in any scoring category.
To determine statistical significance, we performed the G-Test with the ‘DescTools’ package of R Studio (version 1.0.44). The package is loaded with ‘library(DescTools)’. To compare two strains a and b with the respective number of neurons in the neuronal scoring category 1-4, the matrix ‘mat’ was defined by mat = cbind(c(1a,2a,3a,4a), c(1b,2b,3b,4b)). A G-Test on this matrix was performed with ‘GTest(mat)’. The result is given as a P-value.
Code for example above:
> library(DescTools)
> mat = cbind (c(12,2,18,86), c(20,5,28,54))
> GTest(mat)
Output for example above:
Log likelihood ratio (G-test) test of independence without correction
data: mat
G = 12.382, X-squared df = 3, p-value = 0.006182
Notes
- To compare results between strains, it is best practice to test them always in parallel as there will be variations between the assays.
- To test drug response in C. elegans of different life cycle stages, expose a mixed population of animals to 6-OHDA and transfer C. elegans of distinct stages to separate scoring plates using a platinum wire.
- If working with a strain that exhibits a swimming defect (for example unc mutants), synchronize L1 stage animals without starvation by washing off a plate of mixed stage animals and passing them through a 5 μm nylon net filter which will retain animals of all other stages except for L1.
- Aim to use the same lot of 6-OHDA for all experiments as efficiencies between batches can vary. Also keep in mind that 6-OHDA might oxidize over time after opening, so older batches might become less efficient over time.
- The ascorbic acid solution and the 6-OHDA solution must be prepared freshly immediately before treating the animals to avoid oxidization of the compounds.
- Prepare at least 100 μl of each ascorbic acid solution and 6-OHDA solution, as otherwise the amounts will be too small to weigh them out accurately.
- The solutions should appear transparent until addition of M9 buffer. Pink solutions indicate oxidation of 6-OHDA, which does not cause neurodegeneration.
- A higher number of animals can be treated in screw cap tubes and a higher total volume, e.g., in preparation for a qRT-PCR. Synchronized L1 for large approaches can be obtained by starvation of a big plate (keeping the starvation time as short as possible) or by filtering a big, mixed population of animals with a 5 μm nylon net filter.
- If the protocol is used for the paraquat assay, pipet a limited number of animals (ca. 100) on the scoring plates. Count the total number of animals transferred to express the number of animals reaching L3 stage as a percentage.
Recipes
- 200 mM ascorbic acid solution (100 μl)
Weigh 3.52 mg ascorbic acid (MW = 176.12 g/mol)
Vortex solution thoroughly for approximately 1 min until flakes are dissolved and the solution appears as homogeneous - 10 mM 6-OHDA solution (100 μl)
Weigh 1.03 mg 6-OHDA (MW = 205.64 g/mol)
Vortex solution thoroughly for approximately 1 min until it appears as homogeneous.
Acknowledgments
This protocol was adapted from previous work (Nass et al., 2002; Masoudi et al., 2014) and we thank Neda Masoudi for advice. This work was funded by a Wellcome Trust Programme grant to AG (0909444/Z/09/Z, https://wellcome.ac.uk/funding) and a Parkinson’s UK grant (G0912, https://www.parkinsons.org.uk/research/research-grants), together with infrastructure funding from a Wellcome Trust Strategic award (097045/B/11/Z). We acknowledge the Dundee Imaging Facility, which is supported by the Wellcome Trust Technology Platform award (097945/B/11/Z) and the MRC Next Generation Optical Microscopy award (MR/K015869/1). SLO was supported by a Ph.D. fellowship from the Molecular and Cellular Biology programme funded by the Wellcome Trust and by ISSF funding from the Wellcome Trust. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Competing interests
The authors declare that no competing interests exist.
References
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77(1): 71-94.
- González-Hunt, C. P., Leung, M. C., Bodhicharla, R. K., McKeever, M. G., Arrant, A. E., Margillo, K. M., Ryde, I. T., Cyr, D. D., Kosmaczewski, S. G., Hammarlund, M. and Meyer, J. N. (2014). Exposure to mitochondrial genotoxins and dopaminergic neurodegeneration in Caenorhabditis elegans. PLoS One 9(12): e114459.
- He, F. (2011). Common worm media and buffers. Bio-protocol/Bio101 1: e55.
- LB (Luria-Bertani) liquid medium. (2006). Cold Spring Harb Protoc pdb.rec8141. doi:10.1101/pdb.rec8141.
- Masoudi, N., Ibanez-Cruceyra, P., Offenburger, S. L., Holmes, A. and Gartner, A. (2014). Tetraspanin (TSP-17) protects dopaminergic neurons against 6-OHDA-induced neurodegeneration in C. elegans. PLoS Genet 10(12): e1004767.
- Nass, R., Hall, D. H., Miller, D. M., 3rd and Blakely, R. D. (2002). Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci U S A 99(5): 3264-3269.
- Nass, R., Miller, D. M. and Blakely, R. D. (2001). C. elegans: a novel pharmacogenetic model to study Parkinson's disease. Parkinsonism Relat Disord 7(3): 185-191.
- Offenburger, S. L., Ho, X. Y., Tachie-Menson, T., Coakley, S., Hilliard, M. A. and Gartner, A. (2018a). 6-OHDA-induced dopaminergic neurodegeneration in Caenorhabditis elegans is promoted by the engulfment pathway and inhibited by the transthyretin-related protein TTR-33. PLoS Genet 14(1): e1007125.
- Offenburger, S. L., Jongsma, E. and Gartner, A. (2018b). Mutations in Caenorhabditis elegans neuroligin-like glit-1, the apoptosis pathway and the calcium chaperone crt-1 increase dopaminergic neurodegeneration after 6-OHDA treatment. PLoS Genet 14(1): e1007106.
- Schober, A. (2004). Classic toxin-induced animal models of Parkinson's disease: 6-OHDA and MPTP. Cell Tissue Res 318(1): 215-224.
- Sulston, J., Dew, M. and Brenner, S. (1975). Dopaminergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol 163(2): 215-226.
- Tóth, M. L., Simon, P., Kovacs, A. L. and Vellai, T. (2007). Influence of autophagy genes on ion-channel-dependent neuronal degeneration in Caenorhabditis elegans. J Cell Sci 120(Pt 6): 1134-1141.
- Tucci, M. L., Harrington, A. J., Caldwell, G. A. and Caldwell, K. A. (2011). Modeling dopamine neuron degeneration in Caenorhabditis elegans. In: Manfredi, G. and Kawamata H. (Eds.). Methods in Molecular Biology. Humana Press, 129-148.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Offenburger, S. and Gartner, A. (2018). 6-hydroxydopamine (6-OHDA) Oxidative Stress Assay for Observing Dopaminergic Neuron Loss in Caenorhabditis elegans. Bio-protocol 8(18): e3025. DOI: 10.21769/BioProtoc.3025.
Category
Neuroscience > Development > Morphogenesis
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link