- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Activation of Fibroblast Contractility via Cell-Cell Interactions and Soluble Signals
Published: Vol 8, Iss 18, Sep 20, 2018 DOI: 10.21769/BioProtoc.3021 Views: 9506
Reviewed by: Jia LiSalah BoudjadiYoshihiro Adachi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Safety Profiling of Tumor-targeted T Cell–Bispecific Antibodies with Alveolus Lung- and Colon-on-Chip
S. Jordan Kerns [...] Lauriane Cabon
Jan 5, 2023 3699 Views

Biophysical Analysis of Mechanical Signals in Immotile Cilia of Mouse Embryonic Nodes Using Advanced Microscopic Techniques
Takanobu A. Katoh [...] Hiroshi Hamada
Jul 20, 2023 2091 Views

In Vitro Assay to Examine Osteoclast Resorptive Activity Under Estrogen Withdrawal
Cara Fiorino [...] Rene E. Harrison
Jan 5, 2025 1681 Views
Abstract
The collagen contraction assay is an in vitro, three-dimensional method to determine the factor(s) affecting the contractile behavior of activated cells such as fibroblasts in either physiological or pathological scenarios. The collagen lattices/hydrogels are seeded with fibroblasts to mimic the interactions between these cells and their surrounding extracellular matrix proteins in the connective tissue. This method is an important platform to assess components as potential therapeutic targets to prevent pathologies such as fibrosis, which are manifestations of hyperactivated fibroblasts. We have described a basic version of this collagen contraction assay, which is amenable to customization using different cell types under diverse experimental conditions.
Keywords: Collagen contractionBackground
Tissue contraction and remodeling of the extracellular matrix are essential processes in numerous physiological conditions such as wound healing. Central to these two phenomena are fibroblasts, which not only produce and secrete the extracellular matrix proteins but can also reorganize them through mechanical interactions. Interestingly, these cellular behaviors are often exaggerated in pathological conditions such as fibrosis (Desmoulière et al., 2005), thereby illustrating the need to understand the molecular regulation of these processes. Though it has long been known that collagen, one of the main components of the extracellular matrix, is a major player in tissue contraction (Bell et al., 1979), a thorough understanding of the mechanistic details of this process remain elusive. The study of the contraction of fibroblast populated collagen matrices in vitro has enabled researchers to identify novel players which bring about tissue contraction (Ngo et al., 2006; Su and Chen, 2015). Based on this assay, soluble factors such as TGFβ (Levi-Schaffer et al., 1999) and those from immune cells (Garbuzenko et al., 2002; Zagai et al., 2004) have been identified as major factors affecting fibroblast activity. To a large extent, relative to soluble extracellular signals, the contribution of cell-cell interactions on the contractile ability of fibroblasts has been understudied. Recently, we have shown that an important player regulating fibroblast-mediated contraction is the heterotypic cell-cell interactions with immune cells such as mast cells (Pincha et al., 2018). In this protocol, we provide a method to use this assay to determine the effect of cell-cell interactions on matrix contraction. In doing so, we have elaborated upon previously published protocols (Ngo et al., 2006; Su and Chen, 2015) and provide simple alternatives to setting up the assay and the subsequent analysis of the gels. This protocol can also be modified to recapitulate the native/in vivo environments of diverse cell types to assay their regulation within their “normal” microenvironment.
Materials and Reagents
Note: The materials below can also be substituted for equivalent reagents from other companies. Cell culture media and additives will vary based on the cells to be used for the assay. For our protocol, we have used primary dermal fibroblasts from neonatal mice and the mast cell line MCP-5.
- Cell culture and counting
- 70% ethanol
- Recombinant mouse IL-3 (Thermo Fisher Scientific, GibcoTM, catalog number: PMC0034 )
Note: Lyophilized mIL-3 should be stored at 4 °C. Aliquot and store reconstituted mIL-3 at ≤ -20 °C, use within 12 months of reconstitution. - Dulbecco’s Modified Eagle’s Medium (DMEM) (HiMedia Laboratories, catalog number: AL066A )
Note: Store at 4 °C for up to 12 months. - Fetal Bovine Serum (Thermo Fisher Scientific, GibcoTM, catalog number: 26140095 )
Note: Store at -20 °C for up to 12 months. - Sodium Pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11360070 )
- Non-Essential Amino Acids (Thermo Fisher Scientific, GibcoTM, catalog number: 11140050 )
- RPMI media (Sigma-Aldrich, catalog number: R0883 )
Note: Store at 4 °C. Media is light sensitive, use within 12 months of manufacturing. - Solution of penicillin-streptomycin (10,000 U/ml penicillin G sodium and 10,000 μg/ml streptomycin sulfate, Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
Note: Use within 12 months of manufacturing. - Solution of trypsin, 2.5% 10x stock (HiMedia Laboratories, catalog number: TCL051 )
Note: Use within 12 months of manufacturing. - Trypan Blue stain (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061 )
Note: Use within 3 years of manufacturing. - 1x PBS (see Recipes) (Cold Spring Harbor, doi:10.1101/pdb.rec556)
- Collagen lattice
- 10 μl, 200 μl and 1,000 μl pipette tips (any desired make)
- 0.22-μm syringe-driven filter units (Merck, catalog number: SLGV033RS )
- 10 ml Syringes (Dispo Van)
- 24-well cell culture plates (Eppendorf, catalog number: 0030722116 )
- 15 ml Graduated Centrifuge Tubes, sterilized (Tarsons, catalog number: 546021 )
- 1.5 ml microfuge tubes (Corning, Axygen®, catalog number: MCT-150-C )
Note: Need to be autoclaved/sterilized before use. - Rat tail collagen type 1 (Merck, catalog number: 08-115 )
Note: Store at 4 °C, use within 12 months of manufacturing. - Glacial Acetic acid 99.8% (Sigma-Aldrich, catalog number: 109088 )
Note: Store at 15-30 °C. - NaOH pellets (Sigma-Aldrich, catalog number: S8045 )
Note: Store at 15-30 °C. - Stock collagen solution (3 mg/ml) (see Recipes)
- 1 M NaOH Solution (see Recipes)
- Acetic acid solution (0.1%) (see Recipes)
- Image acquisition/analysis
Equipment
- 10 μl, 200 μl and 1 ml pipettes (any desired make)
- Centrifuge (Eppendorf, model: 5810 R or equivalent)
- Hemocytometer (Sigma-Aldrich, catalog number: Z359629 )
- Incubator (CO2 incubator, Thermo Fisher Scientific, model: FormaTM Steri-CycleTM , or equivalent)
- Microscope (OLYMPUS, model: SZX16 or equivalent)
- Digital Camera 3.2 megapixel (Nikon, model: COOLPIX A10 or equivalent) and stand with camera holder
- Flatbed scanner (Epson, model: V39 or equivalent)
Software
- ImageJ Software v. 1.8.0 (National Institutes of Health, Bethesda, MD, https://imagej.nih.gov/ij/download.html) (Schindelin et al., 2012; Rueden et al., 2017)
Procedure
The collagen contraction assay has been previously described for the purpose of assessing the effects of agonists/inhibitors on the contractile properties of fibroblasts (Ngo et al., 2006; Su and Chen, 2015). It provides an easy method to measure the contractile properties of different cell types. An outline of the entire protocol has been provided below (Figure 1). The use of this assay for investigating fibroblast-mediated contraction during heterotypic cell-cell interactions (Pincha et al., 2018) in various conditions has been highlighted here.
Figure 1. Outline of the collagen contraction assay
- Preparation of collagen for making collagen gels
- Acetic acid solution (0.1%) (see Recipe 1)
- Stock collagen solution (3 mg/ml) (see Recipe 2)
- NaOH titration for collagen gel formation
Note: Different lots of Collagen usually have variations in collagen concentration and pH. It is necessary to perform NaOH titration before using a new batch of collagen to optimize solidification.- Aliquot 0.4 ml of DMEM (or media of choice) in eight 1.5 ml tubes.
Note: It is better to use media from an unopened bottle or a freshly prepared lot. Old media aliquots tend to change pH over time, due to the loss of CO2. - Pre-label eight empty wells of a 24-well dish starting from 1 μl to 8 μl. Any old 24-well dish with empty wells can also be used for the titration step.
- Add 0.2 ml of the 3 mg/ml stock collagen solution (prepared in Step A2) to each media containing tube (prepared in Step B1) and mix well by pipetting (avoid bubble formation). The final collagen concentration in each tube should be 1 mg/ml. Note that the addition of collagen solution (with acidic pH) will make the phenol red containing media turn yellow. Since there are no cells at this stage, the collagen can be added to all the tubes at once.
- Add 1 μl of 1 M NaOH to the first tube and mix this solution well with a 1 ml pipet (set at 500 μl) gently for a few times (avoid making bubbles).
- Immediately take 500 μl of the prepared mixture and add it to the pre-designated well of the 24-well dish.
- Repeat Steps B4-B5 with increasing volumes of 1 M NaOH. i.e., 2-8 μl.
- Allow the mixtures to solidify for approximately 20 min at room temperature.
- Choose the least volume of NaOH required to generate a well-solidified circular gel, where the media is pink in color (Figure 2).
Note: Choosing higher NaOH volumes will cause the collagen gels to become more alkaline than ideal, which might not be the best microenvironment for your cells of interest. It also results in gels with very high rigidity, hampering cell-mediated contraction and thus will give erroneous results.
Figure 2. NaOH titration for collagen gel formation. Images of collagen gels prepared with different volumes of NaOH (1-8 μl sequentially added in wells 1-8) taken with Nikon A10 camera. A well solidified circular gel (marked by black arrow) was obtained with 7 μl of NaOH in well 7 and used for later experiments.
- Aliquot 0.4 ml of DMEM (or media of choice) in eight 1.5 ml tubes.
- Preparation of gels for the collagen contraction assay
The following steps have been optimized for analyzing contraction of collagen gels by fibroblasts when they are interacting with another cell type such as mast cells. The same protocol can be used for experiments using fibroblasts alone.- Trypsinize adherent fibroblasts from culture dishes. Remove the spent media, wash with PBS and incubate for 30 sec-1 min with 2 ml of 0.25% Trypsin solution in PBS (1x Trypsin). Neutralize the Trypsin by adding 2 ml of fresh serum containing medium. Collect the detached cells in this 4 ml media solution and transfer to a 15 ml conical tube. Pellet the cells by centrifugation at 150 x g for 4 min, then aspirate and remove the media. For suspension cultures such as mast cells, transfer cell suspension directly to 15 ml conical tubes and pellet by centrifugation at 100 x g for 4 min, then aspirate and remove the spent media.
- Resuspend cell pellet in 1 ml of the media to be used for the experiment.
Notes:- Resuspend cells in complete media or serum-free media depending upon cell type viability without serum. It is advisable to use serum-free media when cells are viable in no-serum conditions. For example, fibroblasts grown in media containing serum often exhibit a higher basal of contraction compared to fibroblasts grown in serum-free media. However, some cell types such as mast cells are not viable without serum and hence complete media must be used in such experimental setups.
- Gel contraction is directly dependent on serum concentrations so when using serum containing media use a uniform concentration across all wells and treatment conditions.
- Determine the number of viable cells in the resuspended pellet. Count the resuspended cells using a hemocytometer and trypan blue dye (or an equivalent alternative).
Note: Maintenance of uniform cell numbers in the collagen gels is crucial and the numbers (i.e., volume of the cell suspension to be used) must be calculated carefully to obtain optimal contraction. Setting up a preliminary experiment to determine basal contraction using different cell numbers when using only one cell type or different ratios in case of analyzing heterologous interaction is recommended. - The number of primary fibroblasts needed per well can range from 50,000 cells/well to 100,000 cells/well of a 24-well dish. The following calculations are done for making technical triplicates. If only fibroblasts are used in the experiment, resuspend 150,000-300,000 cells in 1,200 μl media in a 15 ml conical tube. In experimental setups involving interaction between two different cell types such as fibroblasts and mast cells, the number of mast cells added should be three times the number of fibroblasts. This ratio has been empirically determined for optimal interactions between mast cells and fibroblasts (Pincha et al., 2018). The final volume of the cell suspension for co-cultures should be brought up to 1,200 μl.
Note: It is advisable to have wells in triplicate for each experimental condition. Prepare the mixture for each set of triplicates in the same conical tube or make individual mixtures for each well. - Keep pipettes ready with 600 μl of 3 mg/ml stock collagen solution and the pre-determined volume of 1 M NaOH for 3 wells (Steps B1-B8) and add them immediately to the conical tube with the cell suspension. The final concentration of collagen in the gel mixture will be 1 mg/ml.
- Quickly mix the solution by gently pipetting until the color is a uniform pink (avoid making bubbles) and add 500 μl of the mixture to each pre-designated well of the 24-well dish. Discard any remaining gel mixture.
Note: Always prepare extra volume of the mixture as it ensures proper mixing and transfer of required volume to all the wells and minimizes bubble formation. Proper mixing of NaOH and collagen is essential for maintaining uniform collagen concentration and solidification throughout the gel. Forceful agitation may dissociate collagen fibrils causing poor solidification of gels. - If preparing mixtures for individual well, use one-third of the volumes listed in Steps C4-C5.
Note: A 48-well dish can also be used after reducing the amounts of all cells/reagents by half. It is not advisable to further reduce the volumes or use smaller wells because of the lack of consistency in gel contraction and difficulty in imaging the gels. - Allow the gels to solidify for 20-30 min at room temperature. If the gels have not solidified by then, it indicates improper mixing or low NaOH concentration and it is advisable to start fresh.
- After the gels have solidified, add 500 μl of media to each well.
- Dissociate gels from the wall of the wells by running the tip of a 10 μl pipette tip very carefully along the edges of gels, taking care not to damage or tear the gels.
Note: Make sure to dissociate the gels from the wells only after addition of media as dissociation is easier when the wells have media in them and prevents damage to the gels. - To ensure that gels have completely detached from the well, swirl the 24-well plate gently.
- For experiments to examine the effect of a factor/compound on contraction, add the substance to the media in each well at the required concentration. It is advisable to have wells as negative and positive controls. For negative controls, add media without any test compound. For positive controls add compounds known to cause contraction in the cell type involved, for example, use TGFβ for fibroblasts.
Note: It is recommended that factors/compounds be mixed with the media prior to addition to the gels ensuring uniform distribution. - Place the plates in a 37 °C incubator at 5% CO2 humidifying conditions and take out for imaging at the pre-determined time points.
Note: To determine if a cell type is giving a very high basal contraction, make gels without cells in 3 wells. Use the dimensions of these gels to compare with those seeded with cells. The uniformity of the gels with no cells is also an indicator of correct experimental setup and we recommend setting it up at least the initial few times.
- Measuring collagen contraction
Collagen gel contraction should be measured over various time points after gel solidification (such as 0 h, 4 h, 12 h, 24 h, 48 h and 72 h) as appropriate for the experimental question.- Take the 24-well dish and wrap with parafilm if removing from sterile conditions for imaging.
- Place it on a stereo microscope with an attached camera and image each well using the minimum zoom and appropriate light settings (Figure 3). As an alternative, use a digital camera fixed to a stand at an appropriate height and image each well sequentially or use a flatbed scanner to scan an image of the entire dish.
Note: Use the same settings for all the timepoint measurements for accurate results. The best resolution images with sharp gel outlines are obtained using a microscope or a digital camera.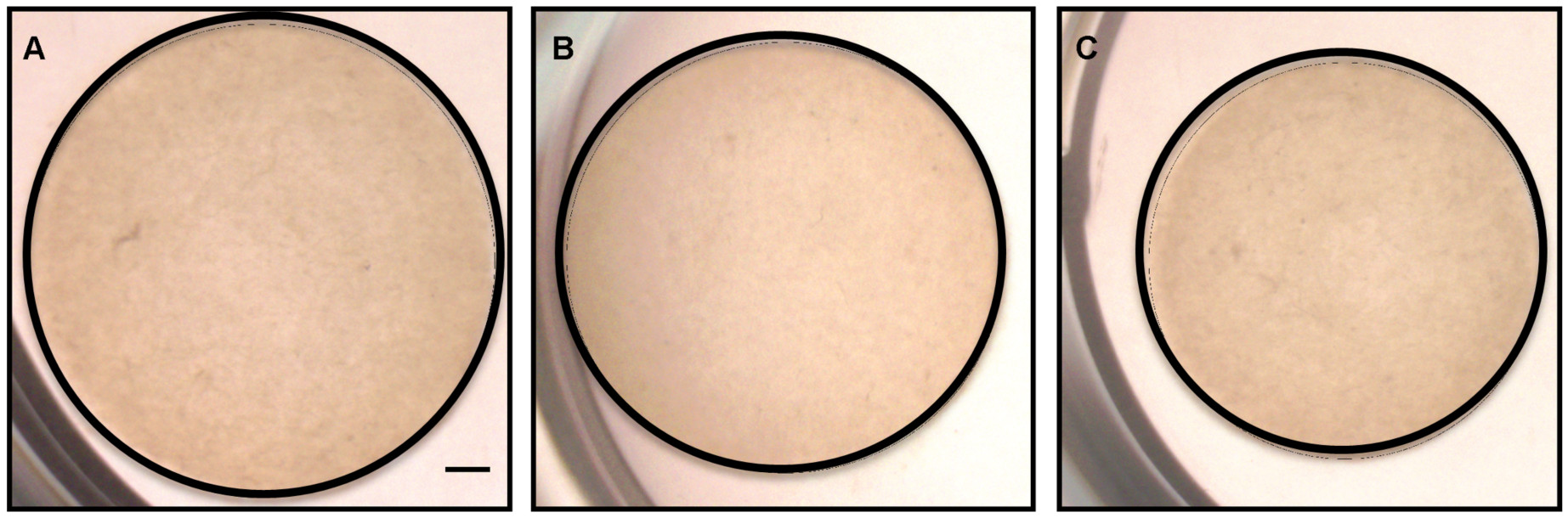
Figure 3. Mast cell-fibroblast co-cultures results in increased collagen contraction at 24 h. Images of collagen gels seeded with: A. No cells; B. Only fibroblasts; C. Mast cells with fibroblasts taken with the Olympus SZX16 microscope at 0.7x magnification (Scale bar = 1 mm). The gels in each panel are outlined with black lines. - Place the dish back into the incubator immediately after imaging (after removing the parafilm and sterilizing the plate with a 70% ethanol wipe) if it is not the end of the experiment.
- When imaging gels at the final time point, better visualization of the gels can be obtained by staining them with crystal violet.
- Remove media from each well very carefully with a pipette. Avoid using vacuum aspirator for this step as it might damage the gels.
- Add 1% crystal violet solution (made in water) to each well and incubate for 5-10 min.
- Pipette out crystal violet carefully, again taking care not to damage the gels.
- Wash the gels with PBS solution till excess crystal violet is removed.
- Remove PBS and image the purple gels (Figure 4).
Notes:- It is easier to image the gels by taking them out from the wells using blunt forceps and placing on a flat surface. However, avoid doing this if gels are too soft as they might break or become distorted.
- The gels can be embedded in paraffin or tissue freezing medium to section and analyze the cell morphology or localization in case of cell-cell interactions.
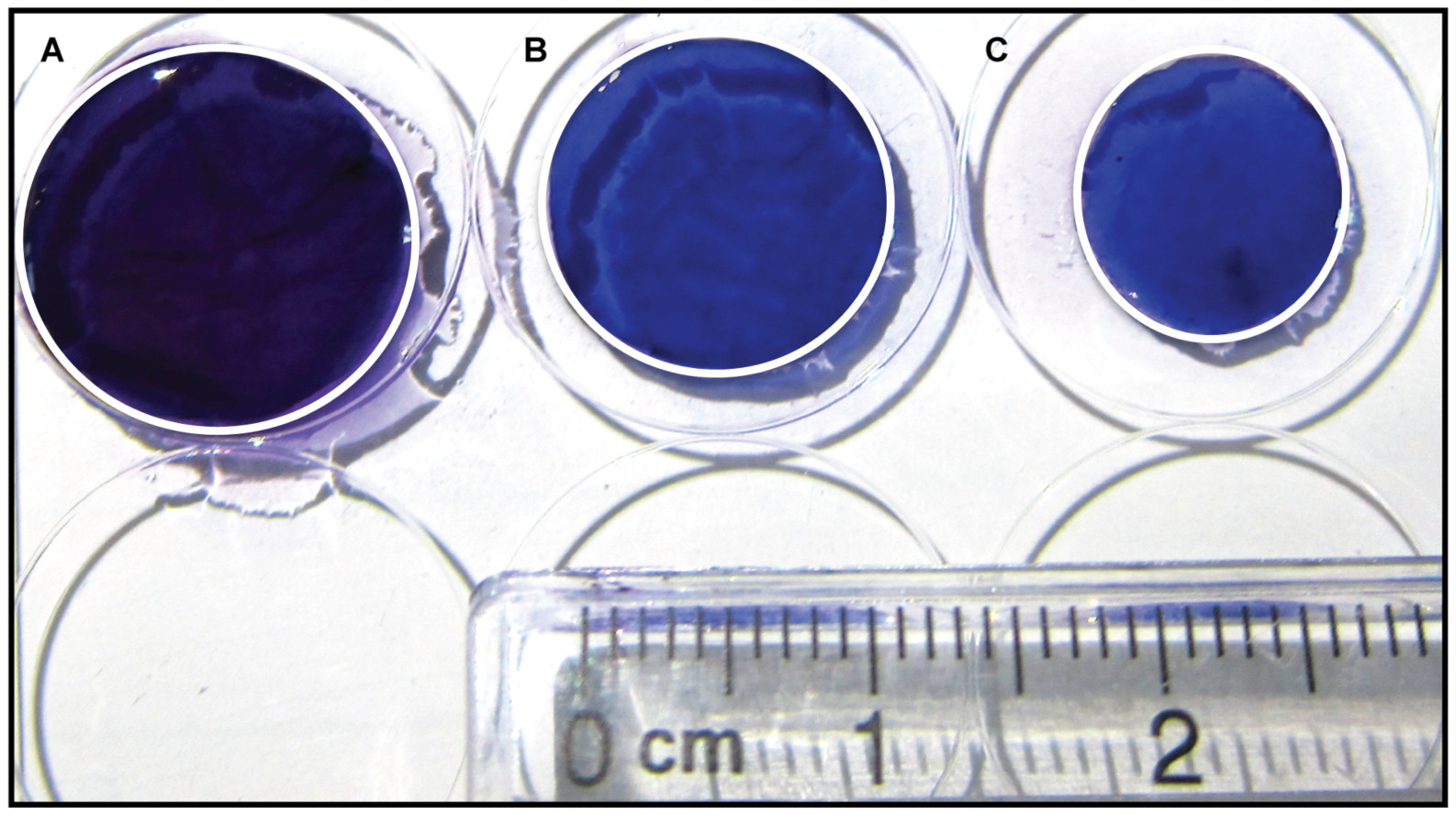
Figure 4. Mast cell-fibroblast co-cultures lead to increased collagen contraction at 72 h. Images of crystal violet stained collagen gels seeded with: A. No cells; B. Only fibroblasts; C. Mast cells with fibroblasts taken with the Nikon A10 camera. The gels have been outlined with white lines.
- Save the images as .tiff files and open with ImageJ (or FIJI). Use these images for calculating collagen gel contraction.
Data analysis
- Open the ImageJ software and in the "Analyze" menu option, check the tick box beside the "area" option in the "Set measurements" option.
- Open each image and draw the outline of the gel using the freehand selection. In case of perfect circular gels, the oval selection can also be used.
Note: Change the brightness or contrast settings of the image to get sharper outlines of the gel for accurate area measurement. - Then for each gel click on "Measure" in the "Analyze" menu (or use the shortcut Ctrl + M) to retrieve the surface area of the gel after drawing the outline.
- Copy the area measurements of all the gels and paste them on an excel sheet for each time point.
- At the end of the measurements, take the mean of triplicates for each condition. The contraction data can be represented in 2 ways using the mean values:
- For each condition, normalize the mean area of each time point with the respective 0 h mean area. Then plot the areas (Y-axis) versus the time points (X-axis) for the respective conditions as lines on the same graph.
- If only the end point measurement is necessary, normalize the mean end point reading of all the conditions to the control at that time point, and show the values as a histogram with the area on the Y-axis and the different conditions on the X-axis.
- Show Inverted values (i.e., 1/area) to represent the increase in collagen contraction. Calculate the standard error of mean using the triplicate values (Figure 5).
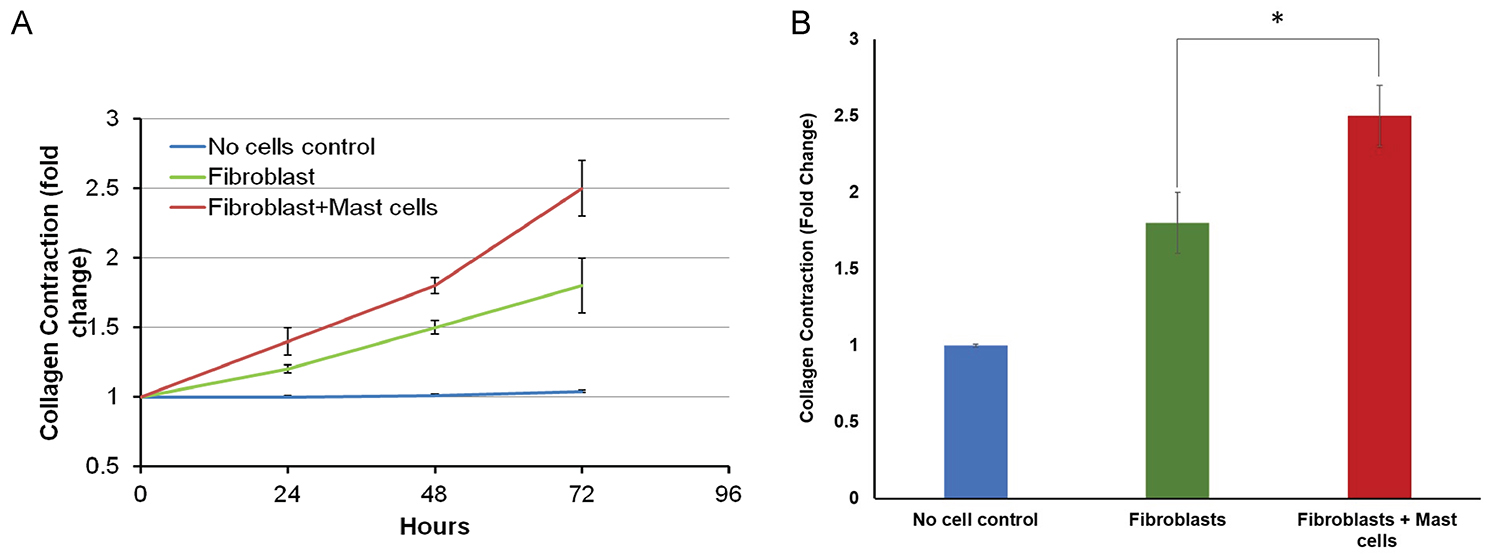
Figure 5. Graphical representation of collagen contraction. A. Collagen gel contraction over 72 h at 24 h time intervals. B. Fold change in collagen gel contraction at 72 h. Statistical analysis has been done using one-way ANOVA followed by Tukey's post-hoc analysis. * = P < 0.05.
Recipes
- Acetic acid solution (0.1%)
- Prepare 25 ml of 0.1% acetic acid by adding 0.025 ml of 100% glacial acetic acid into 24.975 ml deionized water
- Filter sterilize the solution with a 0.2-μm filter and store at 4 °C
- The solution can be stored indefinitely and need not be made fresh every time if kept properly sealed from the external environment
- Stock collagen solution (3 mg/ml)
- Dilute the rat tail type 1 collagen in 0.1% acetic acid (Recipe 1) to make a stock concentration of 3 mg/ml at room temperature
- Ensure that the collagen has dissolved uniformly
- Making small volumes enough for 3-4 experiments ensures uniform solubilization of the collagen
- Store the stock after proper sealing at 4 °C until use
- 1 M NaOH solution
- Dissolve 400 mg weighing NaOH pellets in 8 ml distilled water
- After the pellets dissolve, make up the volume to 10 ml with distilled water
- Filter and store the solution in 1 ml aliquots at 15-30 °C in an air-tight plastic container. Use within 3 months of preparation
- 1x PBS (Dulbecco)
Note: The use of CaCl2 and MgCl2 is optional.CaCl2 (anhydrous) 0.10 g/L KCl 0.20 g/L KH2PO4 0.20 g/L MgCl2•6H2O 0.10 g/L NaCl 8.00 g/L NaH2PO4•7H2O 2.16 g/L Adjust pH to 7.4
Acknowledgments
The authors would like to thank the Jamora lab members for critical review of the experimental details and insightful discussions. This protocol has been adapted from the methods published from the labs of G.T Furuta and E. Song (Ngo et al., 2006; Su et al., 2015). This work was supported by grants from the Department of Biotechnology (Government of India) Grant No. (BT/PR8738/AGR/36/770/2013), NIH/NIAMS (Grant 5R01AR053185-03), the American Cancer Society (Grant 115457-RSG-08-164-01-DDC) and a Hellman Faculty Fellowship to CJ. Animal work was partially supported by the National Mouse Research Resource (NaMoR) Grant (BT/PR5981/MED/31/181/2012; 2013-2016) from the DBT. TB is supported by a Council for Scientific and Industrial Research (Government of India) fellowship. RKZ is supported by a Department of Biotechnology (Government of India) fellowship.
Competing interests
The authors declare that no conflicts of interest exist.
References
- Bell, E., Ivarsson, B. and Merrill, C. (1979). Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci U S A 76(3): 1274-1278.
- Desmoulière, A., Chaponnier, C. and Gabbiani, G. (2005). Tissue repair, contraction, and the myofibroblast. Wound Repair Regen 13(1): 7-12.
- Garbuzenko, E., Nagler, A., Pickholtz, D., Gillery, P., Reich, R., Maquart, F. X. and Levi-Schaffer, F. (2002). Human mast cells stimulate fibroblast proliferation, collagen synthesis and lattice contraction: a direct role for mast cells in skin fibrosis. Clin Exp Allergy 32(2): 237-246.
- Levi-Schaffer, F., Garbuzenko, E., Rubin, A., Reich, R., Pickholz, D., Gillery, P., Emonard, H., Nagler, A. and Maquart, F. A. (1999). Human eosinophils regulate human lung- and skin-derived fibroblast properties in vitro: a role for transforming growth factor β (TGF-β). Proc Natl Acad Sci U S A 96(17): 9660-9665.
- Ngo, P., Ramalingam, P., Phillips, J. A. and Furuta, G. T. (2006). Collagen gel contraction assay. Methods Mol Biol 341: 103-109.
- Pincha, N., Hajam, E. Y., Badarinath, K., Batta, S. P. R., Masudi, T., Dey, R., Andreasen, P., Kawakami, T., Samuel, R., George, R., Danda, D., Jacob, P. M. and Jamora, C. (2018). PAI1 mediates fibroblast-mast cell interactions in skin fibrosis. J Clin Invest 128(5): 1807-1819.
- Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T. and Eliceiri, K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18(1): 529.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Su, S. and Chen, J. (2015). Collagen gel contraction assay. Protoc Exch. doi:10.1038/protex.2015.082.
- Zagai, U., Sköld, C. M., Trulson, A., Venge, P. and Lundahl, J. (2004). The effect of eosinophils on collagen gel contraction and implications for tissue remodelling. Clin Exp Immunol 135(3): 427-433.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pincha, N., Saha, D., Bhatt, T., Zirmire, R. K. and Jamora, C. (2018). Activation of Fibroblast Contractility via Cell-Cell Interactions and Soluble Signals. Bio-protocol 8(18): e3021. DOI: 10.21769/BioProtoc.3021.
Category
Developmental Biology > Morphogenesis > Motility
Cell Biology > Cell-based analysis > Extracellular microenvironment
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












