- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Retroviral Capsid Core Stability Assay
Published: Vol 8, Iss 18, Sep 20, 2018 DOI: 10.21769/BioProtoc.3019 Views: 6466
Reviewed by: Vamseedhar RayaproluRavi KantSzu-Ting Chen

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1878 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2458 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3035 Views
Abstract
Structural stability of the capsid core is a critical parameter for the productive infection of a cell by a retrovirus. Compromised stability can lead to premature core disassembly, exposure of replication intermediates to cytosolic nucleic acid sensors that can trigger innate antiviral responses, and failure to integrate the proviral genome into the host DNA. Thus, core stability is a critical feature of viral replicative fitness. While there are several well-described techniques to assess viral capsid core stability, most are generally time and labor intensive. Recently, our group compared the relative stability of murine leukemia virus capsid cores using an in vitro detergent-based approach combined with ultracentrifugation against the popular fate of capsid assay. We found that both methods reached similar conclusions, albeit the first method was a significantly simpler and faster way to assess relative capsid core stability when comparing viral mutants exhibiting differences in core stability.
Keywords: RetrovirusBackground
Retroviruses have evolved replication cycles that excel in circumventing host antiviral responses. One strategy that retroviruses have developed is to shield their replication intermediates from cytosolic nucleic acid sensors such as cGAS, TREX1, IFI203, and DDX41 (Yan et al., 2010; Gao et al., 2013; Lahaye et al., 2013; Stavrou et al., 2015). During replication, retroviruses produce RNA-DNA hybrids and unmethylated double-stranded proviral DNA in the cytosol that are common targets for innate immune sensors (Yan et al., 2010; Gao et al., 2013; Lahaye et al., 2013). The mature retroviral capsid core is constituted by about 1,500 units of the viral capsid protein (CA) that assemble to form a rigid structure that houses two copies of the retroviral RNA genome, the viral reverse-transcriptase, various other host-derived molecules (e.g., miRNAs, proteins, dNTPs), and in some cases, viral accessory proteins (Ganser et al., 1999; Briggs et al., 2004; Cantin et al., 2005; Campbell and Hope, 2015). This structure is permissive to the diffusion of dNTPs, yet it is impermeable to most host proteins (Jacques et al., 2016). In order to successfully and efficiently deliver the proviral DNA associated with the pre-integration complex (PIC) to the nucleus of the cell, the viral core must maintain a certain level of stability. Once it is physically situated proximal to a nuclear pore complex, the PIC may traverse into the nucleus and deliver the proviral DNA. Capsid cores that lack structural integrity or are destabilized by host restriction factors, like TRIM5α, will trigger innate responses and fail to deliver proviral DNA to the nucleus (Sayah et al., 2004; Stremlau et al., 2004 and 2006).
One of the most commonly used methods of analyzing retrovirus capsid core stability is known as the ‘fate of capsid assay’ (Stremlau et al., 2006; Perron et al., 2007; Yang et al., 2014). This assay involves infecting cells and then detecting the amount of intact pelletable capsid cores in the lysates of the infected cells. While this assay can compare relative levels of cores that persist in the cytosol over time, it is quite labor intensive, time-consuming and requires very large quantities of virus. In our hands, the ‘fate of capsid assay’ required the equivalent of 108 Transducing Units (TU–as measured by productive infections) of Moloney Murine Leukemia Virus (M-MLV) which was barely over the limit of detection in our conditions (Renner et al., 2018a). Additionally, this method cannot differentiate endocytosed, non-infectious capsids, from those capable of a productive infection. Other alternatives to this approach include visual tracking of intact fluorescent cores, and the direct assessment of viral uncoating kinetics in a cyclosporine A washout assay (Fricke et al., 2013; Campbell and Hope, 2015).
The protocol presented here is an adaptation of a similar method (Forshey et al., 2002; Aiken, 2009; Shah and Aiken, 2011). It is designed to rapidly assess the relative stability of capsid cores from different viral mutants. In this modified approach, retroviruses are pre-treated with a detergent to strip away the viral envelope, and then the naked capsids are spun through a sucrose gradient with a detergent layer at the top (Renner et al., 2018a). Measuring the amount of intact cores recovered following ultracentrifugation provides an easy way of determining the comparative stability of the cores from different viruses. In the context of our published study, we also directly compared this method with a conventional ‘fate of capsid assay’, which yielded similar results but was much more involved to carry out and required a substantially larger viral input. The retroviral capsid core stability assay described here poses as an easy and technically reproducible alternative to other approaches evaluating capsid stability (Renner et al., 2018a).
Materials and Reagents
- 10 cm culture dishes (Corning, catalog number: 430167 , or equivalent)
- Pasteur pipettes (Fisher Scientific, catalog number: 13-678-20A , or equivalent)
- Microcentrifuge tubes (FroggaBio, catalog number: LMCT1.7B , or equivalent)
- PVDF membrane (Bio-Rad Laboratories, catalog number: 1620177 )
- Sterile 0.45 μm Luer-Lok syringe filters (Pall, catalog number: 4614 , or equivalent)
- Sterile 20 ml syringes with Luer-Lok (BD, catalog number: 302830 , or equivalent)
- Sterile 50 ml conical tubes (FroggaBio, catalog number: TB50-500 , or equivalent)
- Sterile pipette tips (Diamed Advan Tech, catalog numbers: DIATEC520-5376 ; DIATEC520-5876 ; DIATEC520-6501 , or equivalent)
- Polycarbonate tubes and lids (Beckman Coulter, catalog number: 355618 , or equivalent)
- Serological pipettes, 10 ml (Corning, catalog number: 4488 , or equivalent)
- 293T cells (ATCC, catalog number: CRL-3216 )
- NIH 3T3 (ATCC, catalog number: CRL-1658 )
- R187 Hybridoma (ATCC, catalog number: CRL-1912 )
- Anti-eGFP (Takara Bio, catalog number: 632381 )
- Anti-Mouse IgG, HRP conjugated (Cell Signaling, catalog number: 7076S )
- Anti-Rat IgG, HRP conjugated (Merck, catalog number: AP183P )
- Dulbecco's modified Eagle's medium (DMEM) high glucose, with L-glutamine, sodium pyruvate and phenol red (WISENT, catalog number: 319-005-CL , or equivalent)
- ECL (Bio-Rad Laboratories, catalog numbers: 1705060S [ClarityTM] or 1705062S [Clarity MaxTM], or equivalents)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 12483020 , or equivalent)
- HCl, 36.5-38% (Fisher Scientific, catalog number: A144S-500 )
- Hybridoma-SFM (Thermo Fisher Scientific, GibcoTM, catalog number: 12045076 )
- KCl (Fisher Scientific, catalog number: BP366-500 )
- KH2PO4 (Fisher Scientific, catalog number: P285 500 )
- Methanol (for Transfer Buffer, see Recipes) (VWR, catalog number: 56902-543 )
- Milli-Q Water (or equivalent)
- Na2HPO4 (Fisher Scientific, catalog number: S393-3 )
- NaCl (Fisher Scientific, catalog number: BP358-10 )
- NaOH, 10N certified (Fisher Scientific, catalog number: SS255-1 )
- Penicillin/Streptomycin (GE Healthcare, catalog number: SV30010 , or equivalent)
- Polyethylenimine (PEI) (Polysciences, catalog number: 23966-1 , or equivalent)
- Sodium dodecyl sulphate (SDS) (VWR, catalog number: 97064-496 , or equivalent)
- Sucrose (WISENT, catalog number: 800-081-LG , or equivalent)
- Tris-Base (for transfer buffer, see Recipes) (Fisher Scientific, catalog number: BP152-5 )
- Triton X-100 (VWR, catalog number: 97062-208 , or equivalent)
- 10x PBS (see Recipes)
- 1x PBS (see Recipes)
- PBS-T (see Recipes)
- 20% (m/v) sucrose in PBS (see Recipes)
- 2% (v/v) Triton X-100 or 0.2% (m/v) SDS in 5% (m/v) sucrose in PBS (see Recipes)
- Complete DMEM (see Recipes)
- Transfer Buffer (25x) (see Recipes)
Equipment
- Pipettes (Gilson, catalog number: F167700 , or equivalent)
- 0.22 μm Steritop® filters (Merck, catalog number: SCGPT10RE , or equivalent)
- Balance (Fisher Scientific, catalog number: 01-919-358, or equivalent)
Manufacturer: Ohaus, catalog number: 30100606/RM . - Biosafety cabinet (Thermo Fisher Scientific, model: 1323TS , or equivalent)
- Digital Imager (GE Healthcare, model: ImageQuant LAS 4000, catalog number: 28955810 , or equivalent)
- Fridge (4 °C) (Whirlpool, model: WRT148FZDM , or equivalent)
- Hemocytometer (Hausser Scientific, Bright-LineTM, catalog number: 3100 , or equivalent)
- Microscope (Fisher Scientific, catalog number: LMI6PH2 , or equivalent)
Manufacturer: Laxco, catalog number: LMI6PH2 . - Refrigerated table-top centrifuge (Thermo Fisher Scientific, model: 75004524 , or equivalent)
- Tissue culture incubator, humidity, temperature and CO2 regulated (Thermo Fisher Scientific, model: 3110 , or equivalent)
- Type 70Ti Rotor (Beckman Coulter, catalog number: 337922 , or equivalent)
- Ultracentrifuge (Beckman Coulter, model: 969347 , or equivalent)
Procedure
- Refer to Figure 1 for an overview of the two methodologies used in this protocol.
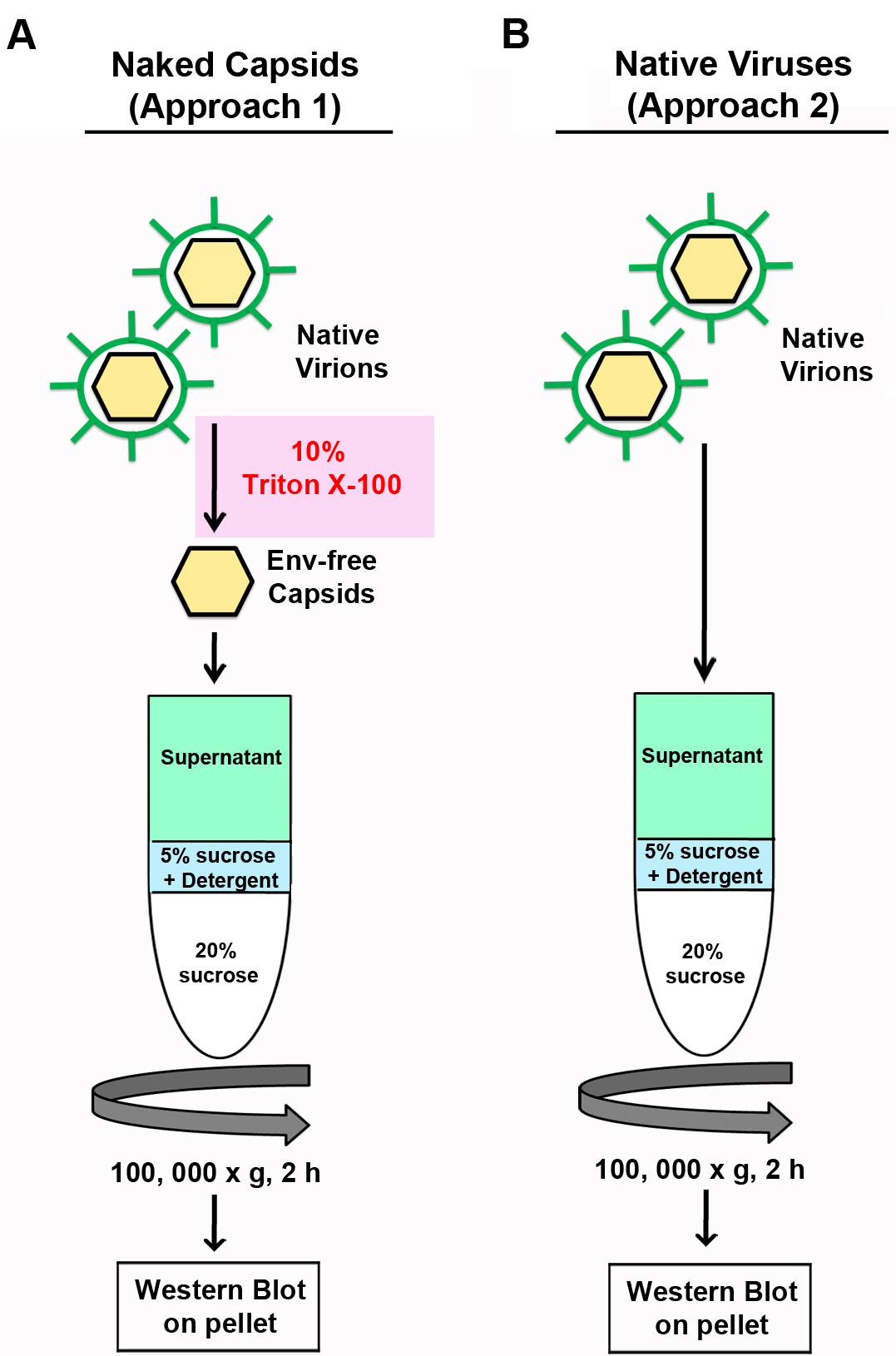
Figure 1. Overview of the Retroviral Capsid Core Stability Assay. Figure adapted from Renner et al., 2018a. A. Approach 1. Retroviruses are stripped of their envelope phospholipid bilayer coat via an initial treatment with detergent. These envelope-deficient (Env-free) capsids are subjected to ultracentrifugation, which briefly exposes them to an additional detergent treatment before entering a detergent-free sucrose solution. The density of this bottom layer containing 20% sucrose allows for efficient pelleting of intact capsids, but is a barrier to pelleting free CA protein and less dense protein complexes. B. Approach 2. A similar approach can be performed without the detergent pre-treatment step on native viruses. In this situation, the viruses will then be slightly more resistant to the detergent layer in the sucrose column. Performing these methods in combination will provide informative results about viral capsid core stability. The pellet is then analyzed by SDS-PAGE using antibodies specific to the CA protein, in this case p30 of M-MLV. - Seed 10 cm dishes with viral producer cells (i.e., viable infected cells that produce representative viral particles) in 8 ml of complete DMEM. For this assay, we typically seed 2 dishes each with 2 x 106 NIH 3T3 cells, chronically infected with MLV. Alternatively, the cells (such as 293T) can be transfected with a virus expressing plasmid (see Notes 1 and 2).
- Allow the cells to propagate and produce virus at 37 °C in 5% CO2 until they reach confluence; this should take approximately 72-96 h. Confluence is determined by lack of surface area remaining on the well's growth surface.
- Before collecting the viral supernatant, pre-cool the ultracentrifuge to 4 °C.
- Collect the viral supernatant (roughly 15 ml) using a serological pipette, and transfer it into a 50 ml conical tube. In these conditions, this is an excess of virus that will allow for multiple attempts at SDS-PAGE analysis if necessary.
- Centrifuge this supernatant for 5 min at 500 x g at room temperature (20-25 °C) to clear cellular debris.
- During this centrifugation step, prepare the appropriate number of syringes and 0.45 μm filters.
- Transfer the cleared supernatant into a syringe and filter it directly into another 50 ml conical tube.
- Approach 1: Add Triton X-100 to a final concentration of 10% (i.e., 1.7 ml Triton X-100 + 15 ml viral supernatant), mix by inversion or pipetting. Incubate at room temperature (20-25 °C) for 20 min (see Note 3).
- Approach 2: Move to Step 9.
- Transfer the solution into a Type 70Ti tube.
- If necessary, top up each tube with media or PBS such that they contain approximately 7.5 ml below the maximum threshold.
- Place a sterile Pasteur pipette into each 70Ti tube, with the thin side immersed in viral media. Placement and use of the Pasteur pipette in this fashion is shown in Figure 2 of a recent publication by our laboratory (Renner et al., 2018b).
- Slowly add the sterile 5% sucrose-detergent buffer of choice, 2 ml of this is sufficient (see Note 4). This allows for a short exposure to detergent before passing through the denser, 20% sucrose solution, which is detergent-free. In our experiments, we typically use 5% sucrose + 2-10% Triton X-100 or 0.2% SDS.
- Using the same Pasteur pipette, slowly add the sterile 20% sucrose solution through the Pasteur pipette so it may form a cushioning layer below the detergent layer (see Note 4). Five milliliters of this solution is sufficient.
- Insert the lids and caps onto the tubes and balance each tube appropriately for ultracentrifugation (within 0.05 g). Sterile media or PBS can be used to adjust the mass of viral samples (see Note 5).
- Cap each tube, ensuring the O-rings and aluminum caps are sealed properly. Insert these tubes appropriately into the Type 70Ti rotor and insert the rotor into the ultracentrifuge (see Note 5).
- Ultracentrifuge these samples with an acceleration and deceleration set to maximum, for 2 h at 4 °C at approximately 100,000 x g.
- Remove the tubes from the rotor, visualize the pellets and circle with a marker. These pellets are rather translucent with a white tinge, as the envelope should have been stripped from them at this stage. As time passes after the centrifugation, these pellets become more difficult to see and may dislodge, so samples need to be processed as fast as possible.
- Gently remove supernatants using a serological pipette and resuspend the pellet in buffer of choice. We typically use 250 µl Laemmli sample buffer for analysis by SDS-PAGE. Resuspend slowly and carefully to avoid the introduction of bubbles.
- Store at -20 °C or -80 °C until ready for analysis.
Data analysis
SDS-PAGE:
Viral capsid core stability can be determined by quantifying essential viral components; loss of viral envelope glycoprotein is expected to occur as the phospholipid bilayer will be dissociated from the capsid. Density of the bottom sucrose layer will govern the stringency and overall yield of this assay. For M-MLV, we targeted the p30 capsid protein (R187, rat monoclonal, 1 µg/ml) and viral envelope glycoprotein Env-eGFP (anti-eGFP, JL-8, mouse monoclonal, 0.2 µg/ml). For the p30 capsid antibody, R187 cells were propagated in the conditions outlined by the ATCC using Hybridoma-SFM. The supernatant may be directly used or purified using Protein A or G resins. The remaining antibodies, including secondaries, were purchased as outlined in the Materials and Reagents section.
This approach and necessary background information are illustrated and interpreted in our recent publication in the Journal of Virology (Renner et al., 2018a). The development of this protocol was to assess the impact on core stability caused by mutations in various viral structural proteins and in an accessory protein called glycosylated Gag (also known as glycogag or gPr80). Figure 2 below depicts the localization of each mutation in the viral proteins. A virus mutant named M-MLV [CTA] specifically depicts the loss of the CTG alternative start codon for gPr80, making this virus deficient in this protein. The other mutants express gPr80 lacking glycosylation sites. These mutations consequently impact the matrix (N113) and CA (N480 and N505) structural proteins.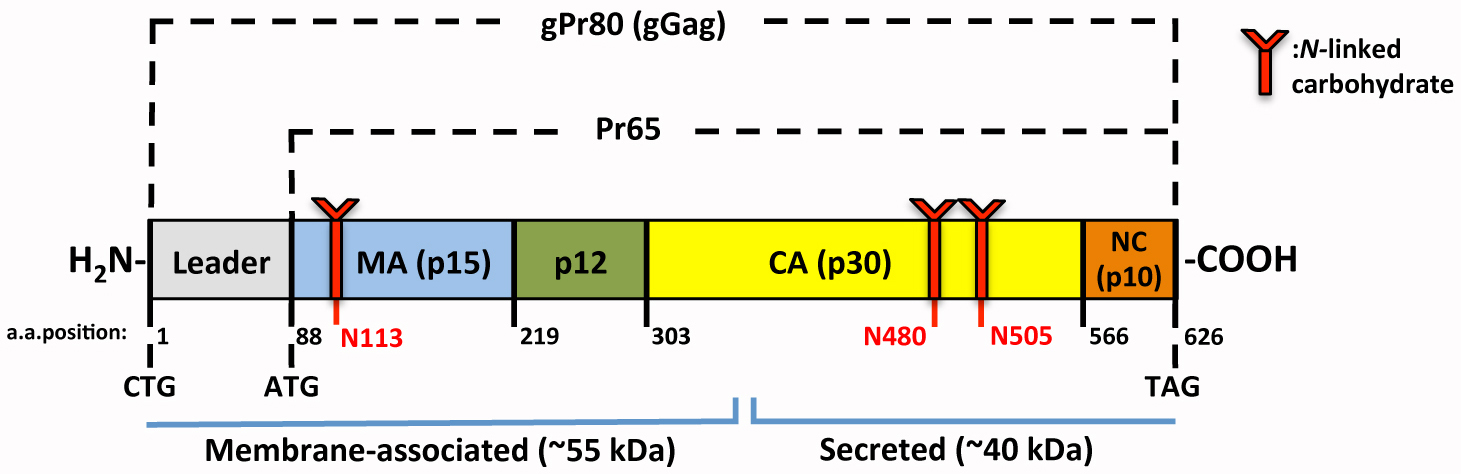
Figure 2. Schematic representation of the glycosylated Gag (gPr80) protein. This figure was adapted from Renner et al., 2018a. Translation of gPr80 is initiated from an alternative start codon, CTG (amino acid (AA) position 1). N-linked glycosylation occurs at AA positions 113, 480 and 505. As illustrated above, these are located in the coding sequences for the Matrix (MA/p15) and Capsid (CA/p30) proteins of Pr65, respectively.
Figure 3 below outlines an example of implementation and data acquired using both approaches 1 and 2 of this protocol. The first lane of each gel shows an untreated control of the viral stock to verify quality of the input virus. Approach 1, which involves pre-treatment of the virus with Triton, completely removes viral envelopes, as depicted by the loss of Env-eGFP staining by Western blot (Figure 3A). This approach clearly shows that loss of gPr80 expression or mutation of N480 results in a detectable reduced capsid core stability (p30 signal), whereas mutating both N113 and N505 do not appear destabilize the capsid core.
Approach 2, which does not involve a pre-treatment with detergent, depicts a gradual loss of the viral envelope with increasing concentrations of detergent, as indicated by diminishing Env-eGFP staining (Figure 3B). This approach illustrates a similar reduction in capsid core stability when comparing the WT M-MLV with the gPr80-deficient virus and the N113Q/N480Q/N505Q mutant. Moreover, Approach 2 reveals more subtle differences between the gPr80-deficient virus and the N113Q/N480Q/N505Q mutant that were not visible in Figure 3A. Chiefly, that [N113Q/N480Q/N505Q] is more stable in the presence of mild detergent than the gPr80-deficient virus. These conclusions were reflected in our direct comparison with the Fate of Capsid Assay in our recent publication (Renner et al., 2018a). 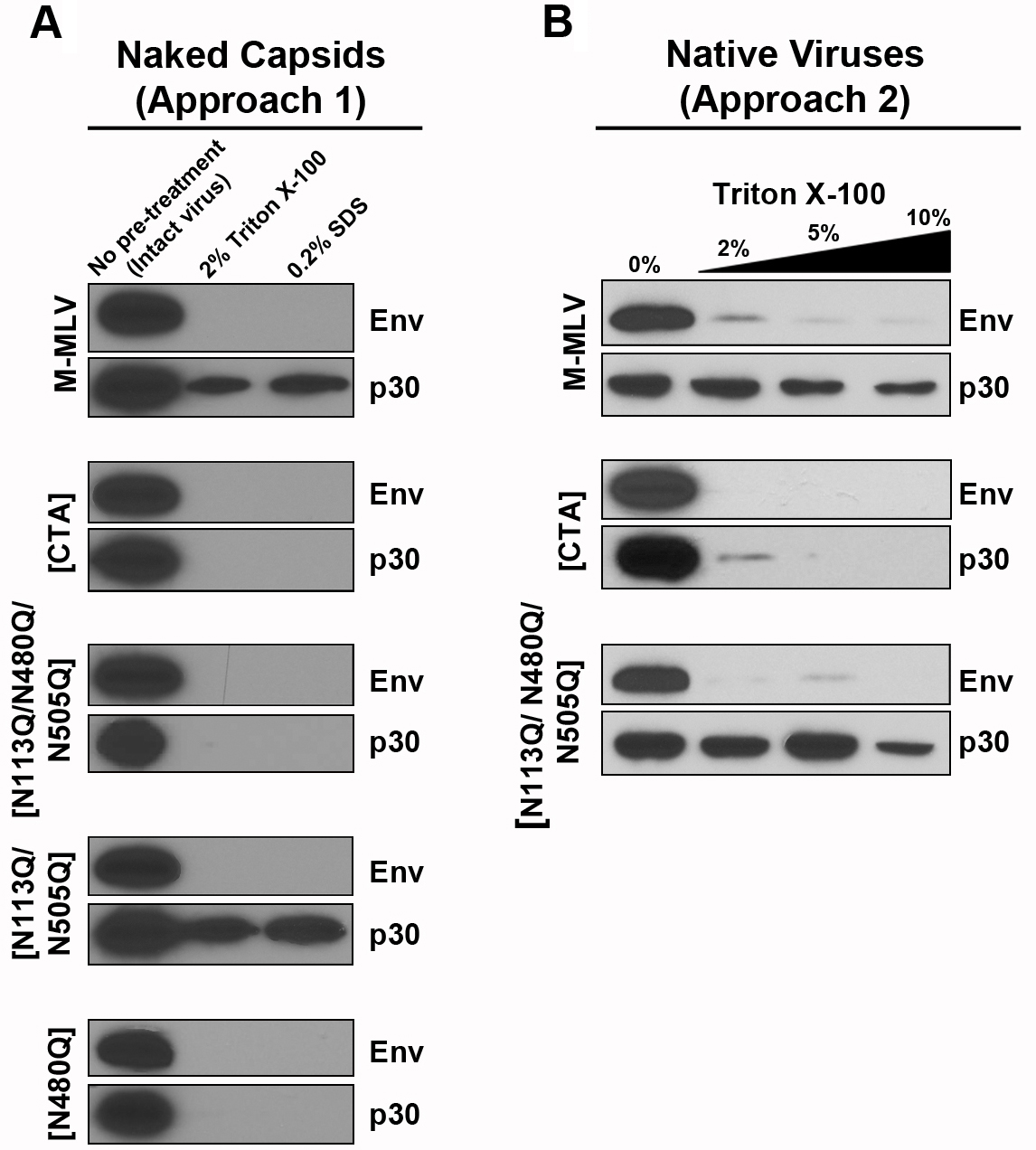
Figure 3. Assessment of viral capsid core stability. This figure was adapted from Renner et al., 2018a. Various M-MLV glycosylated Gag mutants were harvested from cell supernatants as previously described. Intensity of each band is directly proportional to the amount of antibody, and therefore antigen present in the sample; the stronger the band, the more protein of interest is present. A. Viruses were pre-treated with 10% Triton X-100 to remove the envelope (Approach 1). The viruses were then spun through a first layer containing 5% sucrose with either 2% Triton X-100 or 0.2% SDS (Figure 1). Ultracentrifugation pellets were then analyzed by SDS-PAGE. B. Viruses were not pre-treated with any detergent (Approach 2). The first lane represents a sample untreated with detergent, while the next 3 lanes depict treatments with increasing concentrations of detergent within the upper 5% sucrose layer (Figure 1). Absence of an envelope glycoprotein band (Env), but presence of a p30 (capsid) band in SDS-PAGE analysis illustrates envelope-free and intact capsid cores that have survived ultracentrifugation. Reduced or absent p30 staining highlights capsids with decreased structural stability.
Overall, the two technical approaches presented here individually constitute fast, simple and effective ways to monitor capsid core stability of retroviruses. Additionally, by using these two approaches in parallel, it is possible to reproducibly reveal small, but significant, differences in capsid core stability. While Approach 1 challenges the structural stability of naked capsids that have been stripped of their envelope; Approach 2 probes the overall stability of intact, native, viruses challenged with a mild nonionic surfactant. Finally, the results from these two methods were also compared to the well-characterized fate of capsid assay, which yielded similar conclusions in our system, but constitutes a more labor-intensive and time-consuming approach (Renner et al., 2018a).
Notes
- Cell type will vary depending on the virus of choice for this assay. NIH 3T3 cells were chosen as they are easy to work with while being highly permissive to MLV infection and production. If chronically infected producer cells are not a feasible option, transfection of a suitable cell line has also been verified as effective using this protocol. For transfection, follow the published guidelines for your chosen reagents.
- Optimal production time may vary depending on the virus or cell type.
- Pipetting detergent stocks may prove difficult due to their viscosity. Cutting the end of a pipette tip with scissors may help. Alternatively, it may be more accurate to determine the volume of detergent used by mass. Detergent of choice and incubation time may vary depending on virus and experimental conditions. Optimization is recommended with non-denaturing conditions.
- If difficulties are found with the layering of the multiple sucrose layers (i.e., layers are visibly mixing), it is possible to freeze these layers individually with liquid nitrogen prior to adding the viral supernatant. This can be done by immersing the tube containing only the bottom layer of sucrose into a container of liquid nitrogen. Addition of the second layer can be done on top of the frozen bottom layer without fear of mixing. The tube can be immersed once again to freeze both layers in place while the sample is added. This will ensure a proper separation of layers before the centrifugation step has begun. However, just be certain the centrifuge tubes that are being used are compatible with the low temperatures of the liquid nitrogen.
- Be very careful when maneuvering with these tubes, as the sucrose layers can mix with the viral supernatant. It is important that all layers remain distinct and separated.
Recipes
- 10x PBS
80 g/L NaCl
2 g/L KCl
7.63 g/L Na2HPO4
2.4 g/L KH2PO4
Sterilize if to be stored for extended period
Note: We typically make a 10x stock solution of PBS to reduce significance of the error associated with weighing these powders. - 1x PBS
Dilute 10x PBS as necessary to a 1x PBS solution with distilled water
Adjust pH to 7.4 using NaOH or HCl and sterilize
Store anywhere between 4 °C and 25 °C - PBS-T
Add Tween-20 to a final concentration of 0.1% (v/v) (1 ml for each liter) to 1x PBS
Store at room temperature - 20% (m/v) sucrose in PBS
200 g sucrose
100 ml 10x PBS solution
Volume brought to 1 L with sterile H2O
Adjust pH to 7.4 using NaOH or HCl
Filter sterilize
Store in fridge (4 °C) - 2% (v/v) Triton X-100 or 0.2% (m/v) SDS in 5% (m/v) sucrose in PBS
2.5 g sucrose
1 ml Triton X-100 or 0.1 g SDS
5 ml 10x PBS Solution
Volume brought to 50 ml with sterile H2O
Adjust pH to 7.4 using NaOH or HCl
Filter sterilize
Store at room temperature
Note: If a different percentage of Triton X-100 or SDS are required, scale up or down accordingly. - Complete DMEM
(10% FBS, 100 U/ml Penicillin, 100 µg/ml Streptomycin)
DMEM high glucose
50 ml Fetal Bovine Serum
5 ml penicillin/streptomycin solution
Store in fridge (4 °C) - Transfer Buffer (25x)
72.8 g Tris-Base
360 g Glycine
Volume brought to 2 L with sterile H2O
Store this at room temperature
Just before use, dilute to 1x using 20% total volume methanol and the remainder sterile H2O (i.e., 1 L total volume = 200 ml methanol, 40 ml 25x Transfer Buffer and 760 ml sterile H2O).
Acknowledgments
M.-A.L. holds a Canada Research Chair in Molecular Virology and Intrinsic Immunity. This research was supported by a grant from the Canadian Institutes of Health Research (grant 89774) and an Early Researcher Award from the Ontario Ministry of Research and Innovation to M.-A.L. T.M.R. holds a QEII Graduate Scholarship of Ontario.
This protocol was optimized and briefly described in the Journal of Virology (Renner et al., 2018a).
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Aiken, C. (2009). Cell-free assays for HIV-1 uncoating. Methods Mol Biol 485: 41-53.
- Briggs, J. A., Simon, M. N., Gross, I., Krausslich, H. G., Fuller, S. D., Vogt, V. M. and Johnson, M. C. (2004). The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11(7): 672-675.
- Campbell, E. M. and Hope, T. J. (2015). HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13(8): 471-483.
- Cantin, R., Methot, S. and Tremblay, M. J. (2005). Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J Virol 79(11): 6577-6587.
- Forshey, B. M., von Schwedler, U., Sundquist, W. I. and Aiken, C. (2002). Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76(11): 5667-5677.
- Fricke, T., Brandariz-Nunez, A., Wang, X., Smith, A. B., 3rd and Diaz-Griffero, F. (2013). Human cytosolic extracts stabilize the HIV-1 core. J Virol 87(19): 10587-10597.
- Ganser, B. K., Li, S., Klishko, V. Y., Finch, J. T. and Sundquist, W. I. (1999). Assembly and analysis of conical models for the HIV-1 core. Science 283(5398): 80-83.
- Gao, D., Wu, J., Wu, Y. T., Du, F., Aroh, C., Yan, N., Sun, L. and Chen, Z. J. (2013). Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341(6148): 903-906.
- Jacques, D. A., McEwan, W. A., Hilditch, L., Price, A. J., Towers, G. J. and James, L. C. (2016). HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 536(7616): 349-353.
- Lahaye, X., Satoh, T., Gentili, M., Cerboni, S., Conrad, C., Hurbain, I., El Marjou, A., Lacabaratz, C., Lelievre, J. D. and Manel, N. (2013). The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39(6): 1132-1142.
- Perron, M. J., Stremlau, M., Lee, M., Javanbakht, H., Song, B. and Sodroski, J. (2007). The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol 81(5): 2138-2148.
- Renner, T. M., Belanger, K., Lam, C., Gerpe, M. C. R., McBane, J. E. and Langlois, M. A. (2018a). Full-length glycosylated Gag of murine leukemia virus can associate with the viral envelope as a type I integral membrane protein. J Virol 92(6): e01530-17.
- Renner, T. M., Belanger, K., and Langlois, M. A. (2018b). Selective isolation of retroviruses from extracellular vesicles by immunoprecipitation. Bio-protocol 8(17): e3005.
- Sayah, D. M., Sokolskaja, E., Berthoux, L. and Luban, J. (2004). Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430(6999): 569-573.
- Shah, V. B. and Aiken, C. (2011). In vitro uncoating of HIV-1 cores. J Vis Exp(57).
- Stavrou, S., Blouch, K., Kotla, S., Bass, A. and Ross, S.R. (2015). Nucleic acid recognition orchestrates the anti-viral response to retroviruses. Cell Host & Microbe 17(4): 478-488.
- Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. and Sodroski, J. (2004). The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427(6977): 848-853.
- Stremlau, M., Perron, M., Lee, M., Li, Y., Song, B., Javanbakht, H., Diaz-Griffero, F., Anderson, D. J., Sundquist, W. I. and Sodroski, J. (2006). Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci U S A 103(14): 5514-5519.
- Yan, N., Regalado-Magdos, A. D., Stiggelbout, B., Lee-Kirsch, M. A. and Lieberman, J. (2010). The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 11(11): 1005-1013.
- Yang, Y., Luban, J. and Diaz-Griffero, F. (2014). The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol Biol 1087: 29-36.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Renner, T. M., Bélanger, K. and Langlois, M. (2018). Retroviral Capsid Core Stability Assay. Bio-protocol 8(18): e3019. DOI: 10.21769/BioProtoc.3019.
Category
Biochemistry > Protein > Stability
Microbiology > Microbe-host interactions > Virus
Molecular Biology > Protein > Stability
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











