- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Platelet Migration and Bacterial Trapping Assay under Flow
Published: Vol 8, Iss 18, Sep 20, 2018 DOI: 10.21769/BioProtoc.3018 Views: 6913
Reviewed by: Andrea PuharSuprabhat MukherjeeLip Nam LOH

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1335 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1460 Views

An Ex Vivo Lung Histoculture Model for Studying Pulmonary Infection and Immune Response With SARS-CoV-2 as an Example of RNA Virus
Elena V. Maryukhnich [...] Elena J. Vasilieva
Dec 20, 2025 740 Views
Abstract
Blood platelets are critical for hemostasis and thrombosis, but also play diverse roles during immune responses. We have recently reported that platelets migrate at sites of infection in vitro and in vivo. Importantly, platelets use their ability to migrate to collect and bundle fibrin (ogen)-bound bacteria accomplishing efficient intravascular bacterial trapping. Here, we describe a method that allows analyzing platelet migration in vitro, focusing on their ability to collect bacteria and trap bacteria under flow.
Keywords: PlateletsBackground
Platelets are small, anucleate cellular fragments released from megakaryocytes that reside within the bone marrow of mammalian organisms (Machlus and Italiano, 2013). Approximately 750 billion platelets circulate in human blood, constantly scanning the vasculature for damage of the endothelial surface. Upon encountering endothelial injury, platelets are immediately recruited in a well-characterized cascade of events including initial platelet tethering and rolling, followed by platelet activation, adhesion and spreading, eventually leading to fibrin (ogen)-dependent aggregation and subsequent thrombus retraction (Jackson, 2007). Platelet plug formation is a major step in physiological hemostasis, but also in pathological thrombosis after atherosclerotic plaque rupture, triggering myocardial infarction or stroke (Jackson, 2011).
In addition to their well-established role in hemostasis and thrombosis, platelets also evolved diverse immunologic functions (Semple et al., 2011). Being among the first cells recruited to sites of inflammation and infection, platelets play an essential role in initiating intravascular immune responses (Wong et al., 2013). Accordingly, platelets coordinate the recruitment of a variety of immune cells and instruct them with their effector programs (Sreeramkumar et al., 2014). Platelets also have the ability to directly fight pathogens by releasing anti-microbial mediators and/or physically trapping and encapsulating invaders, thus preventing dissemination with the blood flow (Yeaman, 2014).
We recently identified platelet migration as an autonomous platelet function and showed that migration of single platelets provides a mechanism of controlling their interaction with pathogenic bacteria within the microcirculation (Gaertner et al., 2017). Once adhering to immobilized fibrin (ogen), activated platelets use αIIbβ3 integrins to probe the resistance of their local microenvironment. When actomyosin-dependent traction forces overcome substrate resistance platelets eventually polarize and migrate thereby removing and accumulating platelet-bound ligands. As a prominent example, migrating platelets collect and bundle fibrin (ogen)-bound bacteria accomplishing efficient intravascular bacterial trapping. In contrast to phagocytes like neutrophils, platelets behave like “covercytes” that do not internalize collected bacteria but rather accumulate them on their surface within invaginations of the plasma membrane (White, 2005). When exposed to shear stress in vitro platelets strongly bind to piled-up bacteria.
Here we provide a detailed protocol for the isolation of platelets from human blood and microscopic observation of platelet migration and trapping of Escherichia coli (E. coli) under flow.
Materials and Reagents
- Pipette tips 0.1-10 μl (Eppendorf, epT.I.P.S.® Reloads, catalog number: 022491504 )
- Pipette tips 0.5-20 μl (Eppendorf, epT.I.P.S.® Reloads, catalog number: 022491521 )
- Pipette tips 2-200 μl (Eppendorf, epT.I.P.S.® Reloads, catalog number: 022491539 )
- Pipette tips 50-1,000 μl (Eppendorf, epT.I.P.S.® Reloads, catalog number: 022491555 )
- 15 ml Falcon tube (Corning, catalog number: 352096 )
- Tygon 3350 silicone tube (Saint-Gobain Performance Plastics, catalog number: ABW00002 )
- Safety-Multifly-Needle 21G (SARSTEDT, catalog number: 85.1638.935 )
- Membrane adaptor (SARSTEDT, catalog number: 14.1112 )
- 5 ml syringes (BD, Discardit IITM, catalog number: 301285 )
- 50 ml syringes (BD, perfusion, catalog number: 300136 )
- Bottomless 6 channel sticky slide (IBIDI, sticky slides VI0.4, catalog number: 80608 )
- Glass coverslips 24 mm x 24 mm (SCHOTT, NEXTERION®, No. 1.5, catalog number: D263T )
- Cuvette (Eppendorf, catalog number: 952010069 )
- Human blood (taken from male and female healthy donors with the age of 25-40 years old)
- Plasmid ptdTomato (Takara Bio, catalog number: 632531 )
- E. coli strains (DH12S, Invitrogen)
E. coli was transformed with plasmid ptdTomato. TdTomato is encoded within the lac operon and transcription is induced in the presence of isopropylthio-β-galactoside (IPTG) (IPTG binds to the lac repressor and releases lac repressor from the lac operator). The plasmid carries the bla-gene for ampicillin resistance.
Note: E. coli stock was made in LB with 7% dimethyl sulfoxide (v/v) and kept at -80 °C. - Distilled water (ddH2O) (Millipore)
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2438 )
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: D8537 )
- Recombinant human albumin (rHSA) (Sigma-Aldrich, catalog number: A9731 )
Note: Dissolve rHSA in ddH2O to 8% (w/v) solution and store the aliquots at -20 °C. - Prostaglandin I2 sodium salt (PGI2) (Abcam, catalog number: ab120912 )
Note: Dissolve it in DMSO to 10 mg/ml and store the aliquots at -20 °C. - HEPES solution (Sigma-Aldrich, catalog number: H0887 )
- 20% HNO3 (Carl Roth, catalog number: 4337.2 )
- Hexamethyldisilazane (HMDS) (Sigma-Aldrich, catalog number: 440191 )
- Fibrinogen from human plasma (Sigma-Aldrich, catalog number: F3879 )
Note: Dissolve it in 0.9% NaCl to 2 mg/ml and store the aliquots at -20 °C. - Fibrinogen from human plasma, Alexa Fluor 488 conjugate (Thermo Fisher Scientific, Life technology, catalog number: F13191 )
Note: Dissolve it in 0.1 M NaHCO3 (pH 8.3) to 1.5 mg/ml and store the aliquots at -20 °C. - U46619 (Enzo Life Sciences, catalog number: BML-PG023-0001 )
Note: Dissolve it in DMSO to 28.8 mM and store the aliquots at -20 °C. - Adenosine 5′-diphosphate sodium salt (ADP) (Sigma-Aldrich, catalog number: A2754 )
Note: Dissolve it in PBS to 20 mM and store the aliquots at -20 °C. - Thrombin from bovine plasma (Sigma-Aldrich, catalog number: T4648 )
Note: Dissolve it in 0.1% (w/v) bovine serum albumin (BSA) to 100 U/ml and store the aliquots at -20 °C. - LB broth (Sigma-Aldrich, catalog number: L3022 )
Note: 20 g LB powder was dissolved in 1 L ddH2O and autoclaved. - Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I6758 )
Note: Dissolve IPTG in ddH2O to 100 mM and store the aliquots at -20 °C. - Ampicillin sodium salt (Sigma-Aldrich, catalog number: A9518 )
Note: Dissolve ampicillin in ddH2O to 50 mg/ml and store the aliquots at -20 °C. - Sodium citrate tribasic dehydrate (C6H5Na3O7•2H2O) (Sigma-Aldrich, catalog number: S4641 )
- Citric acid monohydrate(C6H8O7•H2O) (Sigma-Aldrich, catalog number: C1909 )
- Glucose (Merck, Calbiochem, catalog number: 346351 )
- Sodium chloride (NaCl) (Carl Roth, catalog number: 3957.3 )
- Sodium bicarbonate (NaHCO3) (Merck, catalog number: 106329 )
- Potassium chloride (KCl) (Carl Roth, catalog number: 6781.3 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C8106 )
Note: CaCl2•2H2O was dissolved in ddH2O to make 100 mM stock solution and stored at 4 °C. - Acid-citrate-dextrose (ACD) buffer, pH 4.7 (see Recipes)
- Modified Tyrode's buffer (see Recipes)
- Fibrin buffer (see Recipes)
Equipment
- Pipette 0.1-2.5 μl (e.g., Eppendorf, Research plus: O18485E)
- Pipette 0.5-10 μl (e.g., Eppendorf, Research plus: O30964E)
- Pipette 10-100 μl (e.g., Eppendorf, Research plus: O30379E)
- Pipette 100-1,000 μl (e.g., Eppendorf, Research plus: O29260E)
- Orbi-Shaker (e.g., Benchmark)
- pH meter (e.g., Mettler Toledo)
- BioPhotometer (e.g., Eppendorf, model: BioPhotometer 6131 )
- Bunsen burner (e.g., Campingaz)
- Centrifuge (e.g., Eppendorf, model: 5804 )
- Cell culture incubator (e.g., Binder)
- Hematology counter (e.g., Horiba Medical)
- KLM spin coater (e.g., Schaefer)
- Inverted phase contrast and fluorescent microscope (e.g., OLYMPUS, model: IX83 )
- Syringe pump (e.g., KD Scientific, model: KDS 100 )
Software
- Live cell imaging software that was connected to the microscope (e.g., CellSense)
- Fiji software [National Institutes of Health (NIH)]
Procedure
- Isolation of washed platelets
- Take human blood from cubital vein by safety multifly-needle on volunteers. Discard the first 1 ml then take another 5 ml into a syringe with 714 μl ACD buffer (see Recipe 1) inside.
Notes:- The volunteers should not take any drugs that affect platelets in the last two weeks.
- Draw blood smoothly to avoid platelets activation.
- Mix the blood with an equal volume of modified Tyrode’s buffer (pH 6.5) (see Recipe 2) in a 15 ml falcon tube.
- Centrifuge the mixture in Step A2 at 70 x g for 35 min without brake at room temperature (RT) to obtain platelet-rich plasma (PRP) retrieved from the upper layer.
- Mingle 3 ml PRP with 7 ml modified Tyrode’s buffer (pH 6.5) in the presence of 0.1% rHSA (w/v) and 100 ng/mg PGI2 in a 15 ml Falcon tube.
- Centrifuge the diluted PRP in Step A4 at 1,200 x g for 10 min at RT.
- Discard the supernatant and suspend the pellet carefully in 1 ml Tyrode’s buffer with pH 6.5.
- Measure the platelet concentration on an automated hematology counter.
- Store the purified platelets at room temperature until use.
Notes:- Platelets cannot be cultured, so they should be prepared daily from freshly drawn blood.
- The whole process of platelets preparation should be gentle enough to prevent platelets activation.
- Take human blood from cubital vein by safety multifly-needle on volunteers. Discard the first 1 ml then take another 5 ml into a syringe with 714 μl ACD buffer (see Recipe 1) inside.
- Bacteria preparation
- Inoculate E. coli from a frozen stock in LB-broth medium with 100 μg/ml Ampicillin.
Note: Conduct E. coli inoculation at the Bunsen burner. - Culture the inoculated E. coli at 37 °C with 260 rpm shaking overnight.
- On the following day, dilute saturated culture of E. coli in LB-broth medium with 1 mM IPTG and 100 μg/ml Ampicillin (1:500 in a volume of 5 ml).
- Culture the diluted E. coli in Step B3 at 37 °C with 260 rpm for 2.5 h to allow growth to mid-log phase.
- Determine the colony forming units (CFU)/ml of cultured E. coli at OD600nm=1 (1, Determine viability of bacteria as a function of OD600nm by plating bacteria and counting colony forming units at various time points with increasing OD600nm. 2, Based on obtained growth curves determine CFUs/ml at OD600nm=1, we obtained 2.8 x 108 CFU/ml at OD600nm = 1.0).
- Measure the E. coli concentration by optical density at OD600nm (expected OD600nm ~0.5-0.6).
- Adjust the cultured E. coli to OD600nm equal to 0.25 in LB medium (0.7 x 108 CFU/ml; 1 ml), centrifuge at 1,200 x g for 5 min and discard the supernatant.
- Suspend the bacteria pellet in equal volume of PBS (1 ml) and keep them on ice under light protection until use.
Note: Perform the bacteria preparation according to the safety regulations of your laboratory.
- Inoculate E. coli from a frozen stock in LB-broth medium with 100 μg/ml Ampicillin.
- Coverslip coating
- Immerse glass coverslips in 20% HNO3 for 1 h on a shaker with the speed set at 60 rpm, then wash with ddH2O four times.
- Soak acid-washed coverslips in ddH2O for another 1 h on the same shaker, and rinse again with ddH2O twice.
- Dry the freshly cleaned coverslips at 90 rps for 10 sec by a KLM spin coater.
- Silanize air-dried coverslips with HMDS at 80 rps for 30 sec using a spin coater to ensure homogeneous silanization.
Note: HMDS is volatile and inflammable, so coat the HMDS in a ventilation hood. - Cut the 6 channel bottomless Ibidi sticky-slide into single channel slides (Figure 1), and mount to the silanized coverslip.

Figure 1. A photo of Ibidi sticky slides. The upper is the 6-channel slide; the lower is the 6- cutted single slides.
- Seed 120 μl prepared E. coli (8.4 x 106 CFUs) in PBS on silanized coverslip in the Ibidi channel at 37 °C for 30 min.
- Coat 120 μl fibrin (see Recipe 4) on the E. coli seeded coverslip for 15 min at RT.
- Wash the fibrin-coated slides 5 times with modified Tyrode’s buffer (pH 7.4; 120 μl) (Figures 2 and 3).
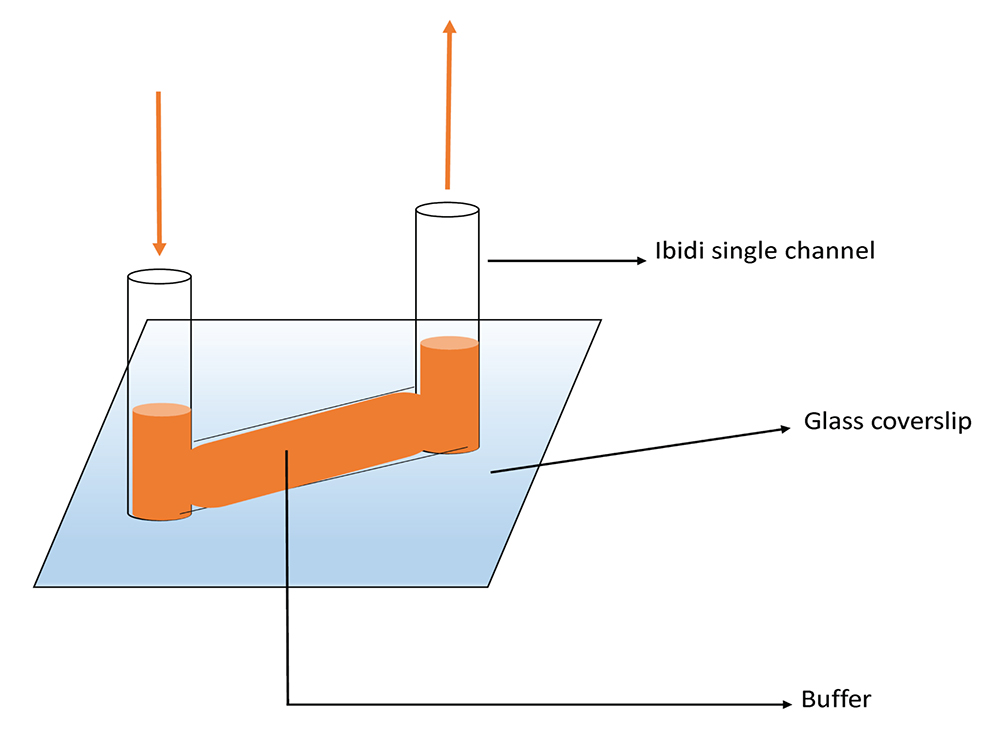
Figure 2. Diagram of the experimental setup. The single Ibidi sticky slide channel is mounted on a silanized glass coverslip. The buffer is pipetted as indicated by the red arrows.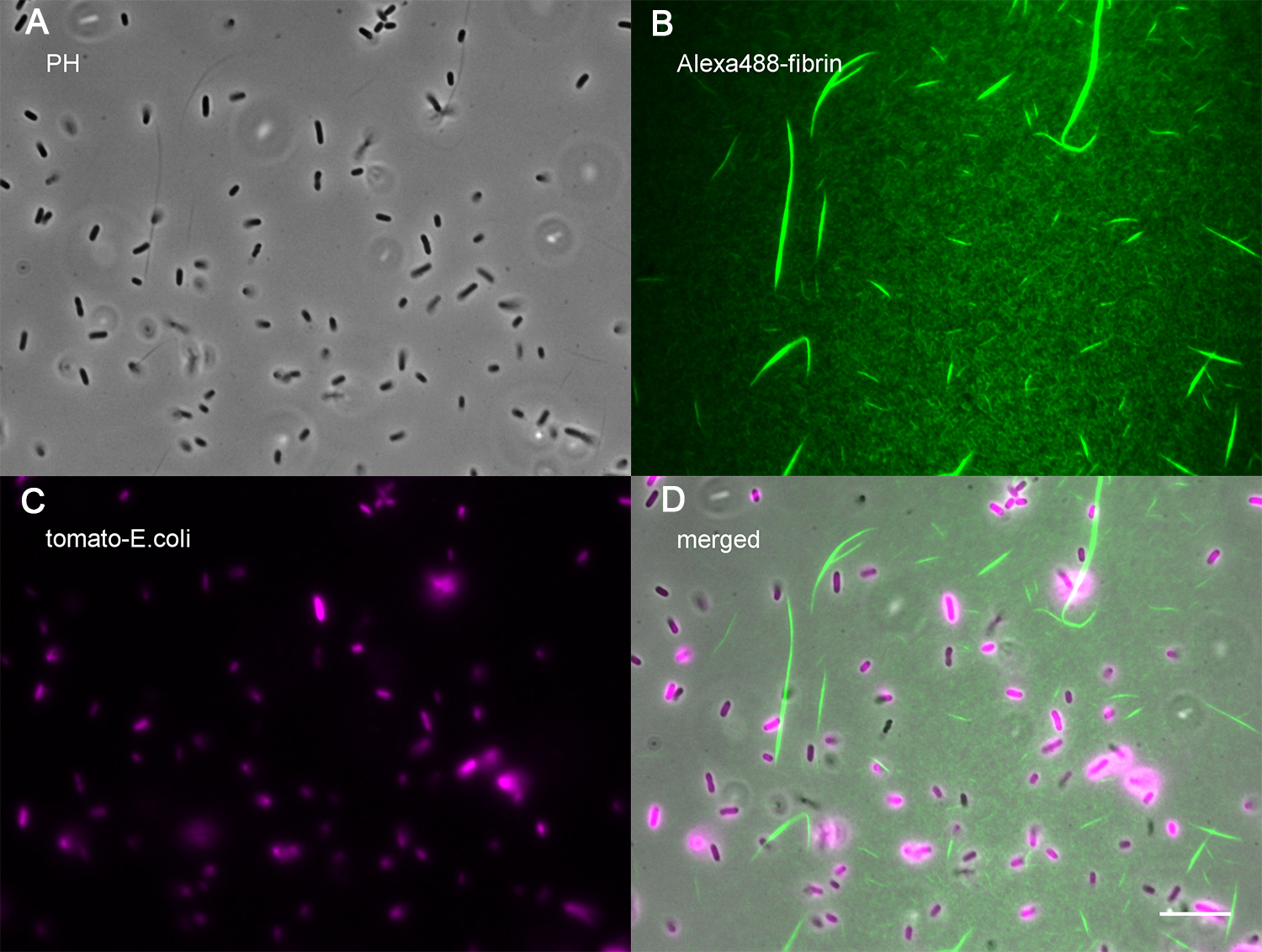
Figure 3. A representative coverslip coated with fibrin and bacteria. Freshly cleaned and silanized glass coverslip was seeded with E. coli and incubated at 37 °C for 30 min, then coated with fibrin for 15 min at RT, and finally washed with Tyrode’s buffer. A. Phase contrast image of the coated channel. B. Fluorescent fibrin coating of the channel. C. Fluorescent E. coli bound to the channel. D. Merged channels. Scale bar = 10 μm.
- Microscopic observation of migrating platelets and bacterial capture
- Supplement modified Tyrode’s buffer pH 7.4 in a total volume of 240 μl with 104/μl human platelets, 1,000 μg/ml rHSA, 200 μM CaCl2, 2 μM U46619 and 4 μM ADP and add to above prepared channels coated with fibrin and live E. coli.
Note: ADP, U46619 and CaCl2 synergistically trigger platelets activation and spreading. Albumin is an anti-adhesive protein that lowers substrate adhesiveness. - Incubate the channel in Step D1 at 37 °C for 1 h and 20 min (Figure 4).
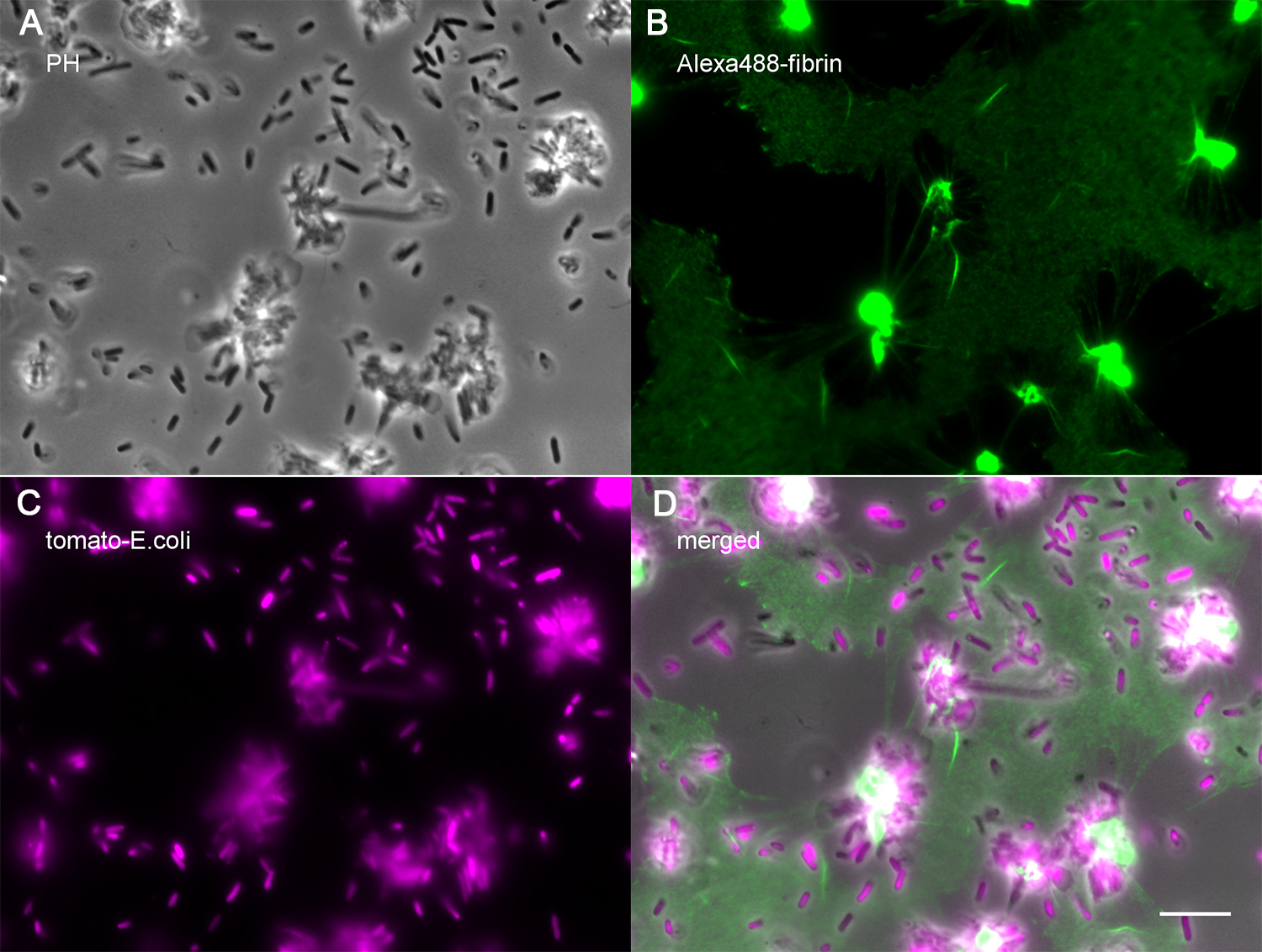
Figure 4. A representative image of migrating platelets collecting fibrin-bound E. coli. Fibrin and live E. coli coated channel was incubated with 104/μl human platelets in the presence of 1,000 μg/ml rHSA, 200 μM CaCl2, 2 μM U46619 and 4 μM ADP in modified Tyrode’s buffer at 37 °C for 1 h and 20 min. Images show phase contrast (A) and epifluorescent micrographs (B and C). The black area in the fibrin image (B) indicates the area cleared by migrating platelets. Scale bar = 10 μm. E. coli accumulated on platelets and bacteria density is reduced in areas where platelets migrated (D). - Assemble the Ibidi channel with a fluid tube connected to the syringe pump and place it on an inverted microscope with a heated stage.
Note: Always carefully rule out air bubbles in the syringe and tube before the perfusion. - Perfuse the channel with pre-warmed (37 °C) modified Tyrode’s buffer (pH 7.4) at 0.5 dyne/cm2 for 1 min to remove non-adhering bacteria and take phase contrast and epifluorescent images.
Note: The flow rate Φ (ml/min) of the syringe pump is calculated by the following formulas provided by the instruction of sticky sides from Ibidi:
τ = η x 97.1 x Φ
τ: Shear stress (dyne/cm2)
η: Dynamical viscosity (dyn s/cm2)
The dynamical viscosity of cell culture medium at 37 °C is ~0.007. - Perfuse the channel with modified Tyrode’s buffer (pH 7.4) at 37 °C at 5 dyne/cm2 for 5 min and take images (Figure 5).
- Image acquisition: Filtercubes (Excitation/Emission): fibrin (Alexa 488) (494/528 nm); E. coli (tdTomato) (590/617 nm).
Note: Deal with the waste of the perfusion buffer that contains bacteria properly according to the safety regulations.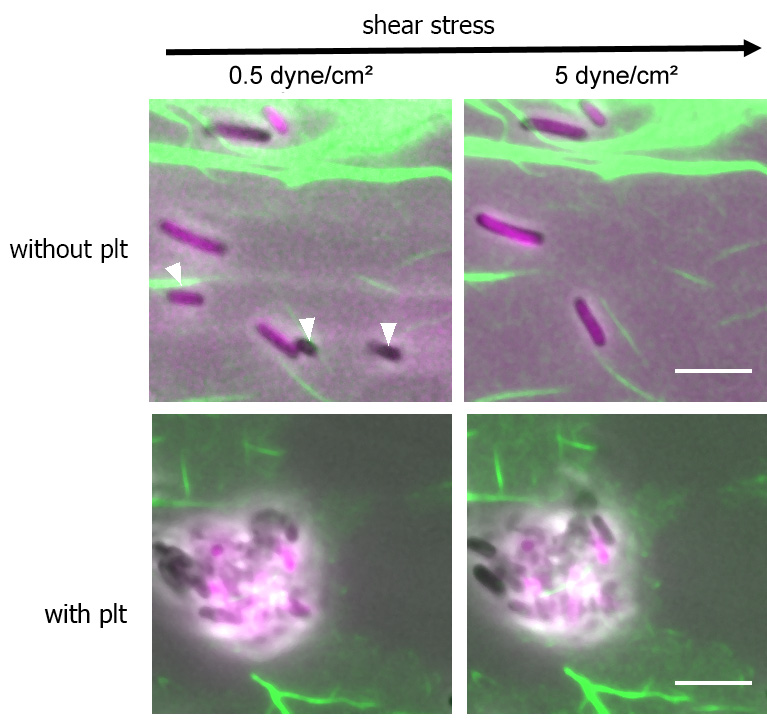
Figure 5. Representative images of bacteria trapped without platelets and with platelets after 0.5 dyne/cm2 and 5 dyne/cm2. The micrographs show the merged phase contrast and fluorescent images. Scale bars = 5 μm.
Note: In the absence of platelets, the number of E. coli (magenta) on the fibrin (green) was reduced after perfusion. The white arrows indicate the E. coli that were flushed away. In contrast, the bacteria bundle collected by migrating platelets is resistant to shear stress.
- Supplement modified Tyrode’s buffer pH 7.4 in a total volume of 240 μl with 104/μl human platelets, 1,000 μg/ml rHSA, 200 μM CaCl2, 2 μM U46619 and 4 μM ADP and add to above prepared channels coated with fibrin and live E. coli.
Data analysis
- Open the acquired images in FIJI software (Schindelin et al., 2012).
- Adjust brightness and contrast of each channel separately to clearly identify bacteria and fibrin fibers, respectively.
- Manually count fibrin-bound bacteria aggregates (in the presence and absence of platelets) before and after perfusion.
- Calculate the fraction of bacteria resistant to perfusion for each experiment by dividing the number of bacteria aggregates after perfusion by the number of bacteria before perfusion. As an example, the bar plot in Figure 6 shows flow resistance of bacterial aggregates in the presence and absence of platelets.
- Test the statistical significance of your results. In the example shown in Figure 6 we repeated experiments 6 times and performed a Wilcox rank sum test to test for statistical significance (R software [R Development Core Team 2011]).
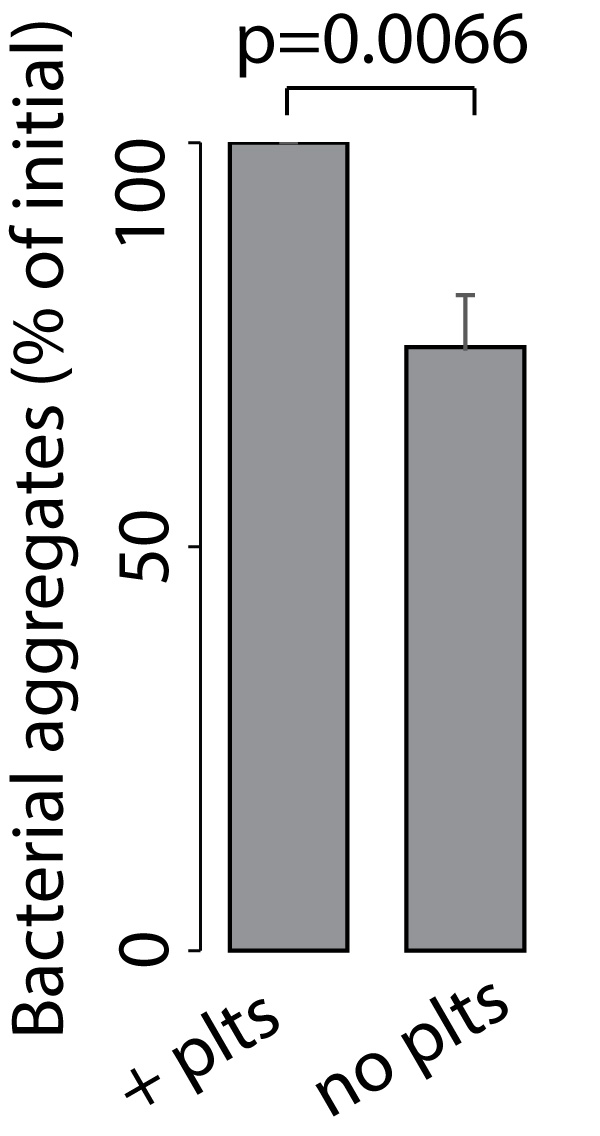
Figure 6. Bar plots showing surface-bound bacterial aggregates after perfusion (5 dyne/cm²) as fraction of initially bound bacteria (in %). Bacteria aggregates bound to platelets are more resistant to shear stress than bacteria aggregates bound to fibrin only. n = 6; Mean ± SEM; statistical significance was tested using the Wilcox rank sum test.
Notes
E. coli (DH 12)-strain used in this protocol requires safety level 1 (S1) laboratories.
Recipes
- ACD buffer, pH 4.7
85 mM Sodium citrate tribasic dehydrate
65 mM citric acid monohydrate
111 mM glucose - Modified Tyrode's buffer
136.9 mM NaCl
12.1 mM NaHCO3
2.6 mM KCl
5.5 mM glucose
10 mM HEPES
Note: Prepare 10x Tyrode's buffer (1.37 mol NaCl, 121 mM NaHCO3, 26 mM KCl) and store at 4 °C. Dilute the stock Tyrode's buffer with ddH2O and add glucose as well as HEPES buffer for daily use. Then adjust the pH to 6.5 for platelets isolation and 7.4 for platelet migration. - Fibrin buffer
Modified Tyrode's buffer (pH 7.4) was supplemented with 300 μg/ml fibrinogen (1:10 mixed with Alexa488 conjugate fibrinogen), 12 mg/ml rHSA, 2 U/ml thrombin, and 1 mM CaCl2
Note: Prepare the fibrin buffer just before use.
Acknowledgments
This protocol was adapted from a previously published study (Gaertner et al., 2017). We thank Michael Lorenz for his excellent assistance in bacteria culture. This work was funded by the DFG SFB 914 (S.M. [B02 and Z01]), the DFG SFB 1123 (S.M. [B06]), the DFG FOR 2033 (S.M. and F.G.), the German Centre for Cardiovascular Research (DZHK) (MHA 1.4VD [S.M.]), FP7 program (project 260309, PRESTIGE [S.M.]), FöFoLe project 947 (F.G.), the Friedrich-Baur-Stiftung project 41/16 (F.G.), Marie Sklodowska Curie Individual Fellowship (EU project 747687, LamelliaActin [F.G.]).
Competing interests
The authors declare that there is no conflict of interest or competing interest regarding the publication of this article.
References
- Gaertner, F., Ahmad, Z., Rosenberger, G., Fan, S., Nicolai, L., Busch, B., Yavuz, G., Luckner, M., Ishikawa-Ankerhold, H., Hennel, R., Benechet, A., Lorenz, M., Chandraratne, S., Schubert, I., Helmer, S., Striednig, B., Stark, K., Janko, M., Böttcher, R. T., Verschoor, A., Leon, C., Gachet, C., Gudermann, T., Mederos, Y. S. M., Pincus, Z., Iannacone, M., Haas, R., Wanner, G., Lauber, K., Sixt, M. and Massberg, S. (2017). Migrating platelets are mechano-scavengers that collect and bundle bacteria. Cell 171(6): 1368-1382 e1323.
- Jackson, S. P. (2007). The growing complexity of platelet aggregation. Blood 109(12): 5087-5095.
- Jackson, S. P. (2011). Arterial thrombosis--insidious, unpredictable and deadly. Nat Med 17(11): 1423-1436.
- Machlus, K. R. and Italiano, J. E., Jr. (2013). The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol 201(6): 785-796.
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Semple, J. W., Italiano, J. E., Jr. and Freedman, J. (2011). Platelets and the immune continuum. Nat Rev Immunol 11(4): 264-274.
- Sreeramkumar, V., Adrover, J. M., Ballesteros, I., Cuartero, M. I., Rossaint, J., Bilbao, I., Nacher, M., Pitaval, C., Radovanovic, I., Fukui, Y., McEver, R. P., Filippi, M. D., Lizasoain, I., Ruiz-Cabello, J., Zarbock, A., Moro, M. A. and Hidalgo, A. (2014). Neutrophils scan for activated platelets to initiate inflammation. Science 346(6214): 1234-1238.
- White, J. G. (2005). Platelets are covercytes, not phagocytes: uptake of bacteria involves channels of the open canalicular system. Platelets 16(2): 121-131.
- Wong, C. H., Jenne, C. N., Petri, B., Chrobok, N. L. and Kubes, P. (2013). Nucleation of platelets with blood-borne pathogens on Kupffer cells precedes other innate immunity and contributes to bacterial clearance. Nat Immunol 14(8): 785-792
- Yeaman, M. R. (2014). Platelets: at the nexus of antimicrobial defence. Nat Rev Microbiol 12(6): 426-437.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fan, S., Lorenz, M., Massberg, S. and Gaertner, F. (2018). Platelet Migration and Bacterial Trapping Assay under Flow. Bio-protocol 8(18): e3018. DOI: 10.21769/BioProtoc.3018.
Category
Immunology > Host defense > Human
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link