- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Selective Isolation of Retroviruses from Extracellular Vesicles by Intact Virion Immunoprecipitation
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.3005 Views: 12160
Reviewed by: Vamseedhar RayaproluSmita NairRajesh Thippeshappa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Respiratory Syncytial Virus with High or Low Content of Defective Viral Particles and Their Purification from Viral Stocks
Yan Sun and Carolina B. López
May 20, 2016 18717 Views

Purification and Identification of Novel Host-derived Factors for Influenza Virus Replication from Human Nuclear Extracts
Kenji Sugiyama and Kyosuke Nagata
Sep 20, 2016 11387 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2167 Views
Abstract
There exists a wide variety of techniques to isolate and purify viral particles from cell culture supernatants. However, these techniques vary greatly in ease of use, purity, yield and impact on viral structural integrity. Most importantly, it is becoming evident that secreted extracellular vesicles (EVs) co-purify with retroviruses using nearly all purification methods due to nearly indistinguishable biophysical characteristics such as size, buoyant density and nucleic acid content. Recently, our group has illustrated a means of isolating intact and highly enriched retroviral virions from EV-containing cell supernatants using an immunoprecipitation approach targeting the viral envelope glycoprotein of the Moloney Murine Leukemia Virus (Renner et al., 2018). This technique, that we call intact virion immunoprecipitation (IVIP), enabled us to characterize the accessibility of epitopes on the surface of these retroviruses and assess the orientation of the virus-encoded integral membrane protein Glycogag (gPr80) in the viral envelope. Proper implementation of this protocol enables fast, simple and reproducible preparations of intact and highly purified retroviral particles devoid of detectable EV contaminants.
Keywords: Retrovirus purificationBackground
Widely used approaches for isolating retroviruses, such as the Human Immunodeficiency Virus (HIV) and Murine Leukemia Virus (MLV), include precipitation, chromatography, ultrafiltration, ultracentrifugation, as well as various other means of particle separation (Reviewed in Nestola et al., 2015). While each technique has its specific advantages, drawbacks and limitations, a common concern for all methods is the co-purification of cell secreted extracellular vesicles (EVs).
EVs constitute a heterogeneous population of membrane-derived vesicles secreted by all cell types (Yanez-Mo et al., 2015). There are strikingly similar biophysical and biochemical characteristics between retroviruses and EVs (Table 1), especially with the small 50-150 nm vesicles secreted through the endosomal pathway, better known as exosomes (Reviewed by Nolte-'t Hoen et al., 2016). Some retroviruses, such as HIV and MLVs, can also share with exosomes the pathways of biogenesis and egress through the endocytic system (Orenstein et al., 1988; Raposo et al., 2002; Houzet et al., 2006; Sandrin and Cosset, 2006; Akers et al., 2013; Madison and Okeoma, 2015; Martin et al., 2016; Nolte-'t Hoen et al., 2016). This imparts inherent biochemical composition similarities between the two types of particles, which extend to their cargo (e.g., proteins, mRNAs, miRNAs) and host-derived surface membrane proteins and antigens (e.g., CD9, CD63, CD81), which inevitably increases difficulties in telling them apart (Eckwahl et al., 2016; Nolte-'t Hoen et al., 2016; Telesnitsky and Wolin, 2016). Given such similarities, selective isolation of retroviruses requires a unique identifying marker to confidently discriminate them from exosomes and EVs in general.
Table 1. Retroviruses and EVs are nearly indistinguishable by their biochemical and biophysical characteristics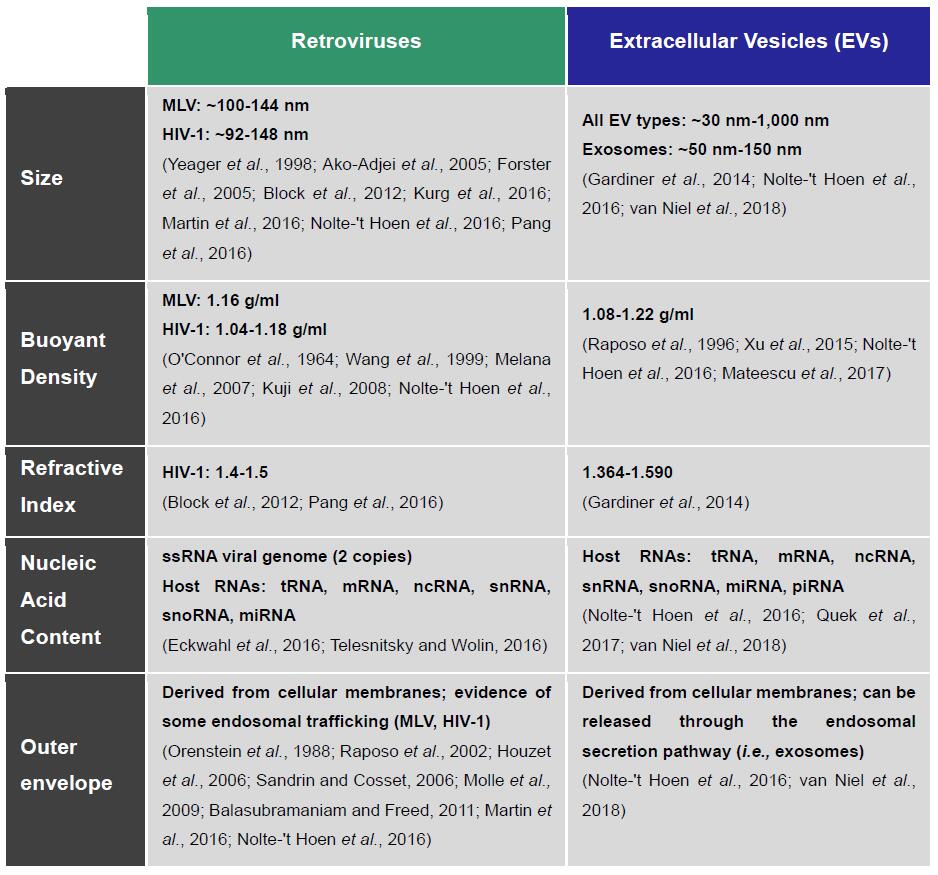
Nanoscale flow cytometry (NFC), also called flow virometry or NanoFACS, is an optimization of flow cytometry techniques, sample preparations and hardware for the analysis of particles smaller than 200 nm, which is the average detection limit of most commercial flow cytometers (Tang et al., 2016 and 2017; Lippe, 2018). This technology is especially useful for the immune phenotypic profiling of markers on the surface of viruses and EVs. By using this approach, we previously determined that a fluorescently tagged viral envelope glycoprotein (Env-eGFP) of the Moloney MLV (M-MLV) was almost exclusively expressed on the surface of these virus particles, and thereby constituted a very reliable selection marker (Figure 1) (Tang et al., 2017).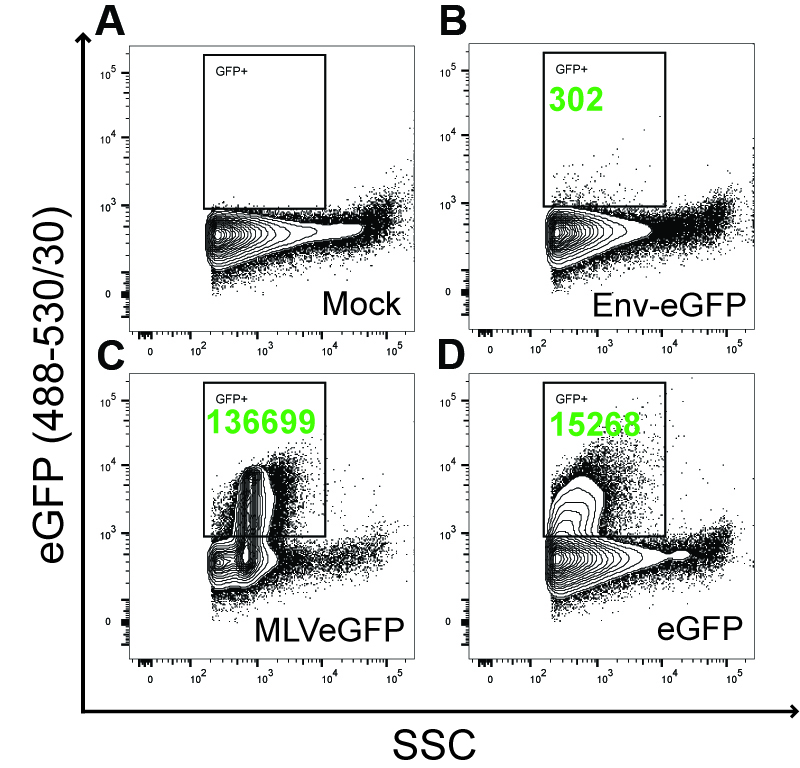
Figure 1. Env-eGFP represents a selection marker for identifying retroviruses by nanoscale flow cytometry. This figure has been adapted from Tang et al. (2017). 293T cells were mock transfected with an empty plasmid (A), with Env-eGFP (B) or eGFP (D) expression plasmids, or with the Env-eGFP expressing M-MLV viral plasmid (C). The M-MLV used in this study contained an eGFP reporter inserted into the proline-rich region of the extracellular domain of the envelope glycoprotein (Sliva et al., 2004). Supernatants were 450 nm-syringe filtered prior to NFC analysis. Transfection efficiency was monitored by eGFP expression in the transfected cells and was similar in each relevant condition (data not shown). For NFC analysis, particles were detected by triggering off of side-scattered light (SSC). A square gate was set above background in the Mock sample where eGFP+ events are expected (A). Side and top boundaries of this gate were determined by the limits of the eGFP+ events in the MLVeGFP sample (C). Numbers in green represent eGFP+ particles detected and enumerated in the gate during a fixed acquisition time window, which was the same for all samples analyzed. For NFC analysis, SSC is more sensitive than forward scattered (FSC) light to detect particles smaller than 200nm on our instrument (Tang et al., 2016). The results show that, in our system, the membrane-expressed Env-eGFP does not substantially associate with EVs (B). However, cytosolic eGFP is incorporated as cargo inside EVs (D). Env-eGFP is highly enriched only on the surface of viruses (C).
The protocol described here was specifically developed to study an enigmatic virus-encoded integral membrane protein called Glycogag (or gPr80) inserted in the envelope of M-MLV (Pillemer et al., 1986; Fujisawa et al., 1997 and 2001; Rosales Gerpe et al., 2015; Renner et al., 2018). Our goal was to assess the incorporation and orientation of full-length gPr80 in the envelope of M-MLV. A major caveat to this particular study was the release of EVs by the infected cells that contaminated our virus samples. This proved to be especially problematic, as we found that the gPr80 protein associated with both EVs and virions (Renner et al., 2018). But given that Env-eGFP was highly enriched on the surface of M-MLV virions and poorly incorporated on EVs (Figure 1) (Tang et al., 2017), we thus developed an intact virion immunoprecipitation (IVIP) assay designed to specifically isolate structurally intact viral particles expressing Env-eGFP on their surface. Using this approach, we successfully identified the orientation of gPr80 as a Type-I integral membrane protein on virions but as a Type-II integral membrane protein on EVs that are devoid of Env-eGFP (Renner et al., 2018). In conclusion, IVIP has the ability to selectively isolate and discriminate retroviruses from EVs with minimal physical manipulation and without compromising the structural integrity of either particle type.
Materials and Reagents
- μ-Columns (Miltenyi Biotec, catalog number: 130-042-701 )
- Microcentrifuge tubes (FroggaBio, catalog number: LMCT1.7B , or equivalent)
- Pasteur pipettes (Fisher Scientific, catalog number: 13-678-20A , or equivalent)
- PVDF membrane (Bio-Rad Laboratories, catalog number: 1620177 )
- Serological pipettes, 10 ml (Corning, catalog number: 4488 , or equivalent)
- Sterile 20 ml syringes with Luer-Lok (BD, catalog number: 302830 , or equivalent)
- Sterile 450 nm Luer-Lok syringe filters (Pall, catalog number: 4614 , or equivalent)
- Sterile 50 ml conical tubes (FroggaBio, catalog number: TB50-500 , or equivalent)
- Sterile pipette tips (Diamed, DIATEC, catalog numbers: DIATEC520-5376 , DIATEC520-5876 , DIATEC520-6501 , or equivalent)
- HEK 293T cells (ATCC, catalog number: CRL-3216 )
- R187 Hybridoma (ATCC, catalog number: CRL-1912 )
- μMACS GFP Isolation Kit (Miltenyi Biotec, catalog number: 130-091-125 )
- 10 cm culture dishes (Corning, catalog number: 430167 , or equivalent)
- 220 nm Steritop filters (Merck, catalog number: SCGPT10RE , or equivalent)
- Anti-eGFP (Takara Bio, Clontech, catalog number: 632381 )
- Anti-Flag, HRP conjugated (Sigma-Aldrich, catalog number: A8592-1MG )
- Anti-Mouse IgG, HRP conjugated (Cell Signaling Technology, catalog number: 7076S )
- Anti-Rabbit IgG, HRP conjugated (Abcam, catalog number: ab6721 )
- Anti-Rat IgG, HRP conjugated (Sigma-Aldrich, catalog number: AP183P )
- Anti-V5 (Merck, catalog number: AB3792 )
- Dulbecco's modified Eagle’s medium (DMEM) high glucose, with L-glutamine, sodium pyruvate and phenol red (WISENT, catalog number: 319-005-CL , or equivalent)
- Dynabeads M270-epoxy (Thermo Fisher Scientific, catalog number: 14321D )
- ECL Substrates, i.e.:
Clarity Western ECL Substrate (Bio-Rad Laboratories, catalog number: 1705060S , or equivalent)
ClarityMax Western ECL Substrate (Bio-Rad Laboratories, catalog number: 1705062S , or equivalent) - Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 12483020 , or equivalent)
- Glycine (Fisher Scientific, catalog number: BP381-5 )
- HCl (36.5-38%) (Fisher Scientific, catalog number: A144S-500 )
- Hybridoma-SFM (Thermo Fisher Scientific, GibcoTM, catalog number: 12045076 )
- KCl (Fisher Scientific, catalog number: BP366-500 )
- KH2PO4 (Fisher Scientific, catalog number: P285-500 )
- Methanol (VWR, catalog number: 56902-543 )
- Milli-Q Water
- Na2HPO4 (Fisher Scientific, catalog number: S393-3 )
- NaCl (Fisher Scientific, catalog number: BP358-10 )
- NaOH, 10N certified (Fisher Scientific, catalog number: SS255-1 )
- NuPAGETM 4-12% Bis-Tris Gel (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0335BOX )
- NuPAGETM MOPS SDS Running Buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0001 )
- Penicillin-Streptomycin (GE Healthcare, catalog number: SV30010 , or equivalent)
- Polyethylenimine (PEI) (Polysciences, catalog number: 23966-1 , or equivalent)
- Sucrose (WISENT, catalog number: 800-081-LG , or equivalent)
- Tris Base (Fisher Scientific, catalog number: BP152-5 )
- TweenTM 20 (Fisher Scientific, catalog number: BP337-100 )
- PBS (10x) (see Recipes)
- 20% sucrose in PBS (see Recipes)
- Complete DMEM (see Recipes)
- Tris-Glycine Transfer Buffer (25x) (see Recipes)
Note: We used VWR as a distributor for Pall and Corning products.
Equipment
- μMACS Separator (Miltenyi Biotec, catalog number: 130-042-602 )
- 4 °C refrigerator
- Balance (Fisher Scientific, catalog number: 01-919-358, or equivalent)
Manufacturer: OHAUS, catalog number: 30100606/RM . - Biosafety cabinet (Thermo Fisher Scientific, catalog number: 1323TS , or equivalent)
- Digital Imager (GE Healthcare, model: ImageQuant LAS 4000, catalog number: 28955810 , or equivalent)
- Haemocytometer (Hausser Scientific, catalog number: 3100 , or equivalent)
- MACS MultiStand (Miltenyi Biotec, catalog number: 130-042-303 )
- Magnetic stand (Thermo Fisher Scientific, catalog number: 12321D )
- Microscope (Fisher Scientific, catalog number: LMI6PH2, or equivalent)
Manufacturer: Laxco, catalog number: LMI6PH2 . - Pipettes (Gilson, catalog number: F167700 , or equivalent)
- Refrigerated table-top centrifuge (Thermo Fisher Scientific, model: SorvallTM ST 40 , catalog number: 75004524, or equivalent)
- Rocking platform (Maxi Rotator) (Labline Instruments, model: Model 4631 , or equivalent)
- Tissue culture incubator, humidity, temperature and CO2 regulated (Thermo Fisher Scientific, catalog number: 3110 , or equivalent)
- Tube Revolver (Thermo Fisher Scientific, catalog number: 88881002 or equivalent)
- Type 70Ti Rotor (Beckman Coulter, catalog number: 337922 , or equivalent)
- Type 70Ti Tubes (Polycarbonate tubes and lids) (Beckman Coulter, catalog number: 355618 , or equivalent)
- Ultracentrifuge (Beckman Coulter, catalog number: 969347 , or equivalent)
Procedure
- Seed 10 cm dishes with 3 x 106 293T cells in 8 ml of complete DMEM. For this assay, we typically use at least 2 dishes per condition.
- Allow the cells to grow at 37 °C in 5% CO2 until they reach 70-80% confluence, this should take approximately 24 h.
- Transfect viral plasmid(s) of interest. For M-MLV, we typically use 10 μg of plasmid DNA, though this may be optimized based on the virus (Note 1).
- Return the cells to the incubator for virus production for a period of 72 h (Note 2).
- Before collecting the viral supernatant, pre-cool the ultracentrifuge to 4 °C.
- Collect the viral supernatant (roughly 15 ml) using a serological pipette, and transfer it into a 50 ml conical tube.
- Centrifuge this supernatant for 5 min at 500 x g to clear cellular debris.
- During this centrifugation step, prepare the appropriate number of syringes and 450 nm filters.
- Transfer the cleared supernatant into a syringe and filter it directly into a Type 70Ti tube.
- Top up each tube with media or PBS such that they contain approximately 5.5 ml below the maximum threshold.
- Place a sterile Pasteur pipette into each 70Ti tube, with the thin side immersed in viral media. Refer to Figure 2.
- Slowly add the sterile 20% sucrose solution through the Pasteur pipette so it may form a cushioning layer below the viral supernatant. Five milliliters of this solution is sufficient (Note 3). Refer to Figure 2.
- Balance each tube appropriately for ultracentrifugation. Sterile media or PBS can be used to adjust the mass of viral samples.
- Cap each tube, ensuring the O-rings and aluminum caps are sealed properly. Insert these tubes appropriately into the Type 70Ti rotor and insert the rotor into the ultracentrifuge (Note 3).
- Ultracentrifuge these samples with an acceleration and deceleration set to 50%, for 3 h at 4 °C at 100,000 x g (Note 4).
- Remove the tubes from the rotor, visualize the pellets and circle with a marker. As time passes after the centrifugation, these become more difficult to see so you need to proceed as fast as possible. Refer to Figure 2.
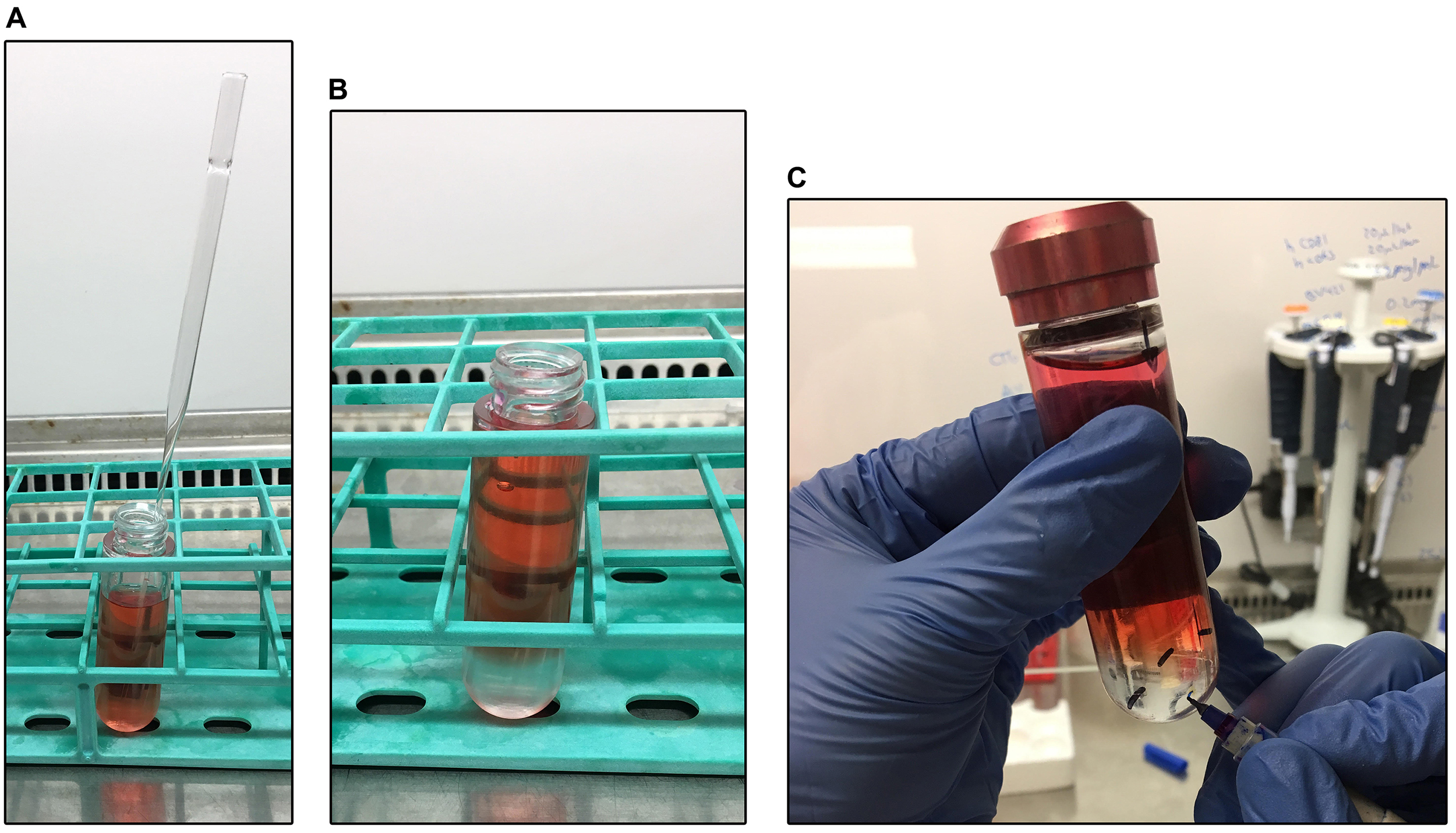
Figure 2. Virus concentration by ultracentrifugation. A Pasteur pipette (A) is used to add a distinct underlayer of sucrose (B). After ultracentrifugation, the pellet is visualized and identified with a marker (C). - Gently remove supernatants using a serological pipette and resuspend the pellet in 1 ml of PBS (Note 4).
- Incubate this concentrated viral sample with an excess of antibody-conjugated beads (we used 30 μl) targeting a surface antigen (i.e., anti-GFP) for 3 h at 4 °C with constant gentle rotation.
- Using the μMACs system (Note 5), ready the μ-columns on the MultiStand.
- Add PBS to prime the column prior to sample addition (Note 6).
- Load the viral sample into a μ-column and allow PBS to flow through the column by gravity. Flow through from this stage can be collected for analysis with the enriched portion. Refer to Figure 3.
- Wash the column with 5x volume of sterile PBS (5 ml for each 1 ml of virus).
- Elute from the column as desired. Miltenyi describes multiple conditions for elution, both denaturing and non-denaturing. In our case, we desired to reduce non-specific elution and maintain viral integrity. To achieve this, remove the column from the magnetic stand, insert it into a microcentrifuge tube and add 250 μl of sterile PBS to the column, which is collected in the tube. The eluate will also contain the magnetic beads, so it will be a translucent brown colour as shown in Figure 3.
- Samples can be stored short-term (< 1 day) at 4 °C, and long-term (weeks) at -80 °C.
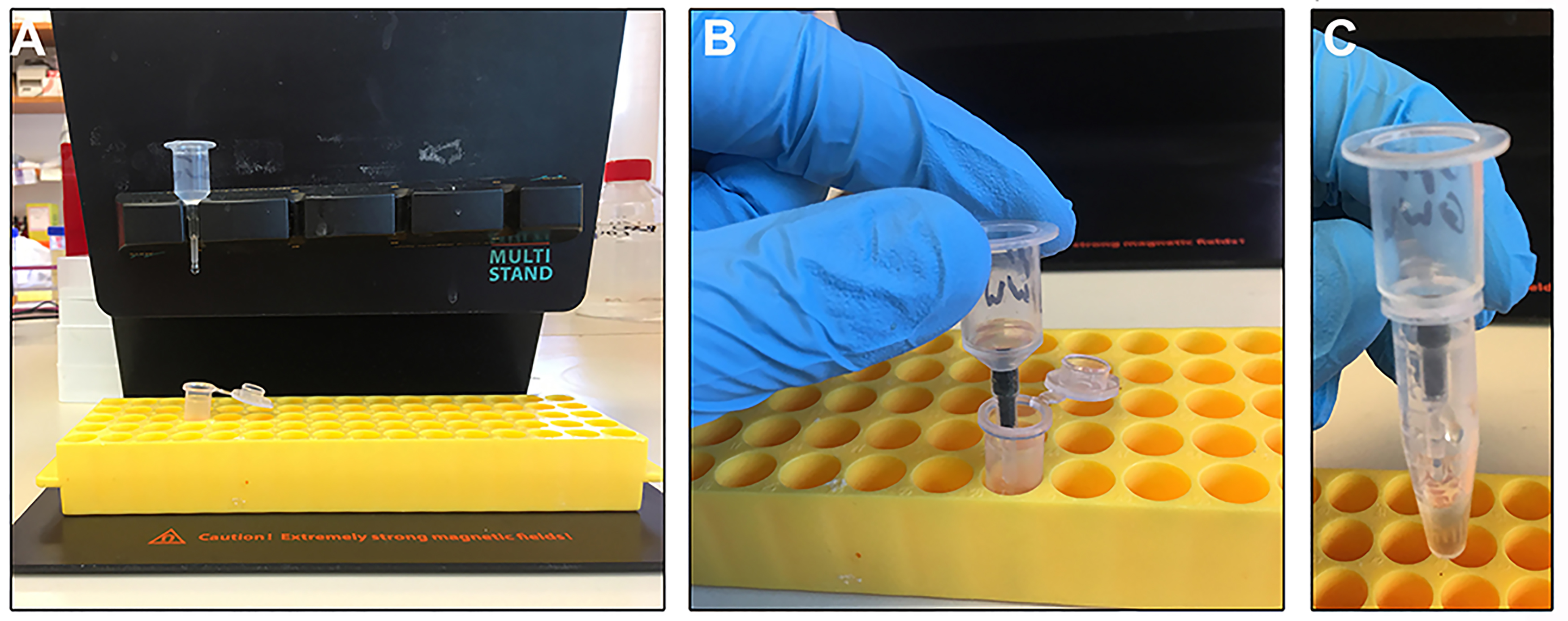
Figure 3. μMACS isolation of intact viruses and gentle elution. A μ-column is securely fit into the separator on the magnetic stand and a microcentrifuge tube can be placed below to collect flow through (A). Removal of the μ-column from the magnetic field (B) will enable mobility of the magnetic beads in any desired elution buffer. The beads will also elute from the column, giving the eluate a translucent brown colour (C).
Data analysis
- SDS-PAGE analysis: Sample purity can be determined by quantifying integral viral components. Proteomic techniques, such as SDS-PAGE or ELISA, are recommended as this is a protein interaction-based isolation. However, viral genome quantifications are a suitable alternative to assess total virus isolation efficiency. In our study, M-MLV was probed for the p30 capsid protein (R187, rat monoclonal, 1μg/ml), the viral envelope glycoprotein Env-eGFP (anti-eGFP, JL-8, mouse monoclonal, 0.2μg/ml) and recombinant gPr80 (gPr93FV contains an N-proximal Flag-tag and C-terminal V5-tag, shifting its observed size from 80 kDa to 93 kDa; anti-V5, rabbit polyclonal, 0.2μg/ml). For the p30 capsid antibody, R187 cells were grown according to the conditions outlined by the ATCC using Hybridoma-SFM. The supernatant can either be used directly or purified using a Protein A or G resin. All other antibodies were purchased from the suppliers indicated in the Materials and Reagents section.
For best resolution, we used NuPAGE 4% to 12% gradient gels in MOPS running buffer at 200 V for approximately 45-55 min. Transfer was done to a PVDF membrane using a Tris-Glycine Transfer buffer at 100 V for approximately 80 min. Blocking was done using 5% milk in PBS-T for 1 h at room temperature. Blocking, washing and antibody staining steps are best performed on a rocking platform. We achieved optimal results by performing primary antibody stainings overnight at 4 °C in blocking buffer, while secondary antibodies (conjugated to HRP) can be incubated at room temperature for 1 h at the proper dilutions in blocking buffer (anti-mouse IgG secondary 1:5,000; anti-rabbit IgG secondary at 0.1 μg/ml; anti-rat IgG secondary at 0.05 μg/ml). Detection was done using an ECL of appropriate intensity (i.e., Bio-Rad, Clarity, 1705060S or ClarityMax, 1705062S, or equivalents) and analyzed on a Digital Imager (GE LifeSciences, ImageQuant LAS 4000, or equivalent). It is good practice to analyze the unprocessed sample (input), flow through (unbound) and eluate (IP) of the immunoprecipitation using multiple antibodies targeting viral components as a way to monitor the performance of the IVIP assay. - Interpretation of results: The IVIP protocol described here was developed to characterize the differential association of the retroviral integral membrane protein glycogag (gPr80) with intact EVs and viruses, along with determining its membrane topology at the surface of M-MLV virions. To help characterize the association of gPr80 with virions and EVs, we developed gPr93FV, a recombinant gPr80 construct harboring N-proximal Flag and C-terminal V5 epitope tags. These features enabled us to determine the orientation of gPr93FV in the outer envelope of EVs and M-MLV by the way it interacts with antibodies directed against each epitope tag. Given that we previously demonstrated that the viral envelope glycoprotein, Env (or more specifically, Env-eGFP), is a selection marker abundantly found on the surface of M-MLV and rarely detected on EVs (Tang et al., 2017), it was therefore possible to distinguish EVs (unbound fraction) and viruses (bound fraction) following the eGFP-targeting IVIP protocol described above.
The Input, Unbound and IP fractions were probed using three antibodies targeting viral proteins. Figure 4A shows that capsid (p30) was present in the Unbound, but enriched in the IP fraction as expected. Env-eGFP, the capture antigen, was present in the IP fraction but not in the unbound fraction, indicating a successful and pure intact virus preparation (Figure 4B). Using a V5 antibody, gPr93FV was detected in both the Unbound and IP fractions (Figure 4C). These data along with other experiments from our study enabled us to conclude that gPr80 can be inserted in the M-MLV envelope as a Type-I integral membrane protein in an NexoCcyto orientation, as its N terminus is accessible to the V5 antibody on the surface of intact virus particles. Development of this protocol has also lead to the identification of subpopulations of secreted particles (viruses, EVs, VLPs) that independently and differentially incorporate viral proteins, and would otherwise be indistinguishable by conventional methods that denature the structural integrity of such particles prior to SDS-PAGE analysis (Renner et al., 2018).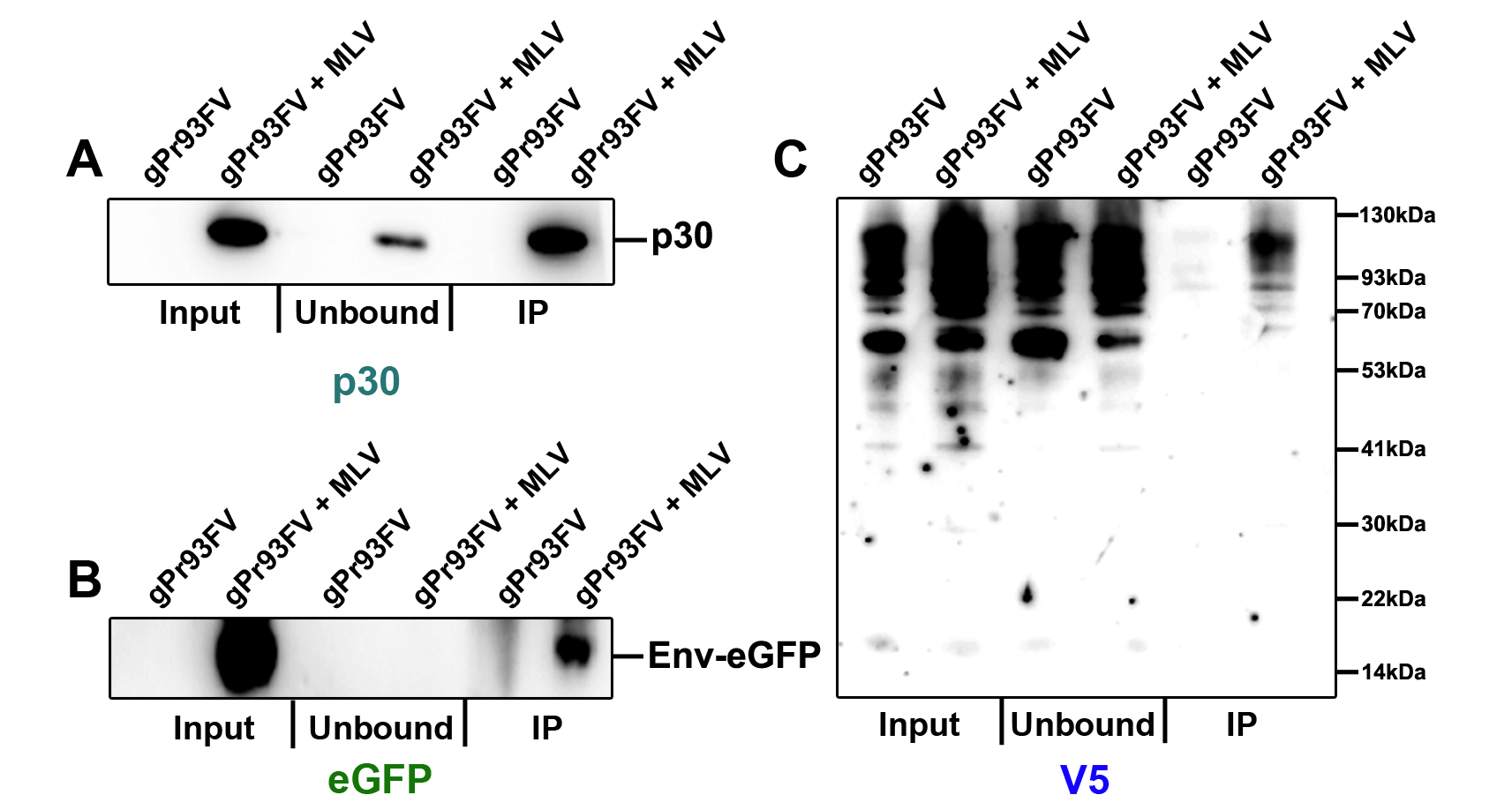
Figure 4. Assessment of the purity and viral constituents of IVIP samples by SDS-PAGE. This figure has been adapted from Renner et al. (2018). M-MLV was co-produced with a Glycosylated Gag expression plasmid (gPr93FV) and isolated according to the IVIP protocol described above. A) Viral capsid (p30), B) viral envelope glycoprotein (eGFP) and C) gPr93FV (V5) were all visualized by SDS-PAGE using NuPAGE gels. The band pattern of gPr93FV is indicative of its multiple glycosylation states. The V5 antibody used visualized a non-specific band at approximately 65 kDa, however it is not a viral component as it is washed away during the IP procedure.
Notes
- Transfection reagents or methods to produce virus from an expression plasmid can vary. We used polyethylenimine (PEI) and the protocol is extensively described in Longo et al. (2013).
- Optimal production time may vary depending on the virus or cell type.
- Be very careful when maneuvering with these tubes, as the sucrose layer can mix with the viral supernatant. It is important that these layers remain distinct and separated.
- Steps 9-17 were carried out using standard methods for M-MLV concentration by ultracentrifugation. Spinning the virus through a sucrose cushion removes small cells debris, cytosolic material and impurities, and typically yields > 90% recovery on input virus infectivity. The pellet generated by ultracentrifugation is expected to be a small, viscous, slightly reddish accumulation at the bottom of the tube (if produced in DMEM with phenol red). Other concentration methods may be more suitable, and gentler, for other types of viruses (Rayaprolu et al., 2018).
- The μMACS system is one option of many which also allows protein G conjugation of any antibody. We have also used Dynabeads M270-epoxy (Thermo Fisher Scientific) in conjunction with the associated magnetic stand (Thermo Fisher Scientific), which allows for the use of any antibody with minimal antibody shedding from the beads. However, magnetic beads are highly recommended, as we find that these often produce reduced levels of non-specific binding when compared to sepharose or agarose beads.
- If gravity flow does not initiate when preparing the column with PBS, a detergent-containing buffer (i.e., PBS + 0.1% TweenTM 20) may be used to prime the column. Be sure to remove all traces of this buffer with a larger volume of detergent-free buffer (i.e., PBS), as the detergent will likely impact the integrity of enveloped viruses/vesicles.
Recipes
- PBS (10x)
NaCl 80 g/L
KCl 2 g/L
Na2HPO4 7.63 g/L
KH2PO4 2.4 g/L
Sterilize by passing through a Steritop 220 nm filter if to be stored for an extended period. Storage at room temperature
Dilute as necessary to a 1x PBS solution
Adjust pH to 7.4 using NaOH or HCl and filter sterilize
Store anywhere between 4 °C and 25 °C
Note: We typically make a 10x stock solution of PBS to reduce significance of the error associated with weighing these powders. This should be stored at room temperature; refrigeration may cause precipitation of salts. - 20% sucrose in PBS (m/v)
200 g sucrose
100 ml 10x PBS solution
Volume brought to 1 L with sterile H2O
Adjust pH to 7.4 using NaOH or HCl
Filter sterilize
Store in fridge (4 °C) - Complete DMEM
DMEM high glucose
50 ml Fetal Bovine Serum
5 ml penicillin/streptomycin solution
Store in fridge (4 °C) - Tris-Glycine Transfer buffer (25x)
72.8 g Tris-Base
360 g Glycine
Volume brought to 2 L with sterile H2O
Store this at room temperature
Just before use, dilute to 1x using 20% total volume methanol and the remainder sterile H2O (i.e., 1 L total volume = 200 ml methanol, 40 ml 25x Transfer Buffer and 760 ml sterile H2O).
Acknowledgments
M.-A.L. holds a Canada Research Chair in Molecular Virology and Intrinsic Immunity. This research was supported by a grant from the Canadian Institutes of Health Research (grant 89774) and an Early Researcher Award from the Ontario Ministry of Research and Innovation to M.-A.L. T.M.R. holds a QEII Graduate Scholarship of Ontario. This protocol was developed and briefly described in the Journal of Virology (Renner et al., 2018). Our gratitude goes to Vera A. Tang for helpful discussions and technical support, especially with regards to nanoscale flow cytometry.
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Akers, J. C., Gonda, D., Kim, R., Carter, B. S. and Chen, C. C. (2013). Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol 113(1): 1-11.
- Ako-Adjei, D., Johnson, M. C. and Vogt, V. M. (2005). The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J Virol 79(21): 13463-13472.
- Balasubramaniam, M. and Freed, E. (2011). New insights into HIV assembly and trafficking. Physiuology (Bethesda) Aug 26(4):236-251.
- Block, O., Mitra, A., Novotny, L. and Dykes, C. (2012). A rapid label-free method for quantitation of human immunodeficiency virus Type-1 particles by nanospectroscopy. J Virol Methods 182(1-2): 70-75.
- Eckwahl, M. J., Telesnitsky, A. and Wolin, S. L. (2016). Host RNA packaging by retroviruses: A newly synthesized story. MBio 7(1): e02025-02015.
- Forster, F., Medalia, O., Zauberman, N., Baumeister, W. and Fass, D. (2005). Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci U S A 102(13): 4729-4734.
- Fujisawa, R., McAtee, F. J., Favara, C., Hayes, S. F. and Portis, J. L. (2001). N-terminal cleavage fragment of glycosylated Gag is incorporated into murine oncornavirus particles. J Virol 75(22): 11239-11243.
- Fujisawa, R., McAtee, F. J., Zirbel, J. H. and Portis, J. L. (1997). Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: identification of differences in processing in vitro and in vivo. J Virol 71(7): 5355-5360.
- Gardiner, C., Shaw, M., Hole, P., Smith, J., Tannetta, D., Redman, C. W. and Sargent, I. L. (2014). Measurement of refractive index by nanoparticle tracking analysis reveals heterogeneity in extracellular vesicles. J Extracell Vesicles 3: 25361.
- Houzet, L., Gay, B., Morichaud, Z., Briant, L. and Mougel, M. (2006). Intracellular assembly and budding of the Murine Leukemia Virus in infected cells. Retrovirology 3: 12.
- Kuji, N., Yoshii, T., Hamatani, T., Hanabusa, H., Yoshimura, Y. and Kato, S. (2008). Buoyant density and sedimentation dynamics of HIV-1 in two density-gradient media for semen processing. Fertil Steril 90(5): 1983-1987.
- Kurg, R., Reinsalu, O., Jagur, S., Ounap, K., Vosa, L., Kasvandik, S., Padari, K., Gildemann, K. and Ustav, M. (2016). Biochemical and proteomic characterization of retrovirus Gag based microparticles carrying melanoma antigens. Sci Rep 6: 29425.
- Lippe, R. (2018). Flow virometry: a powerful tool to functionally characterize viruses. J Virol 92(3).
- Longo, P. A., Kavran, J. M., Kim, M. S. and Leahy, D. J. (2013). Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol 529: 227-240.
- Madison, M. N. and Okeoma, C. M. (2015). Exosomes: Implications in HIV-1 Pathogenesis. Viruses 7(7): 4093-4118.
- Martin, J. L., Cao, S., Maldonado, J. O., Zhang, W. and Mansky, L. M. (2016). Distinct particle morphologies revealed through comparative parallel analyses of retrovirus-like particles. J Virol 90(18): 8074-8084.
- Mateescu, B., Kowal, E. J., van Balkom, B. W., Bartel, S., Bhattacharyya, S. N., Buzas, E. I., Buck, A. H., de Candia, P., Chow, F. W., Das, S., Driedonks, T. A., Fernandez-Messina, L., Haderk, F., Hill, A. F., Jones, J. C., Van Keuren-Jensen, K. R., Lai, C. P., Lasser, C., Liegro, I. D., Lunavat, T. R., Lorenowicz, M. J., Maas, S. L., Mager, I., Mittelbrunn, M., Momma, S., Mukherjee, K., Nawaz, M., Pegtel, D. M., Pfaffl, M. W., Schiffelers, R. M., Tahara, H., Thery, C., Tosar, J. P., Wauben, M. H., Witwer, K. W. and Nolte-'t Hoen, E. N. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles 6(1): 1286095.
- Melana, S. M., Nepomnaschy, I., Sakalian, M., Abbott, A., Hasa, J., Holland, J. F. and Pogo, B. G. (2007). Characterization of viral particles isolated from primary cultures of human breast cancer cells. Cancer Res 67(18): 8960-8965.
- Molle, D., Segura-Morales, C., Camus, G., Berliz-Torrent, C., Kjems, J., Basyk, E., Bertrand, E. (2009). Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J Biol Chem Jul 17; 284(29): 19727-43
- Nestola, P., Peixoto, C., Silva, R. R., Alves, P. M., Mota, J. P. and Carrondo, M. J. (2015). Improved virus purification processes for vaccines and gene therapy. Biotechnol Bioeng 112(5): 843-857.
- Nolte-'t Hoen, E., Cremer, T., Gallo, R. C. and Margolis, L. B. (2016). Extracellular vesicles and viruses: Are they close relatives? Proc Natl Acad Sci U S A 113(33): 9155-9161.
- O'Connor, T. E., Rauscher, F. J. and Zeigel, R. F. (1964). Density gradient centrifugation of a murine leukemia virus. Science 144(3622): 1144-1147.
- Orenstein, J. M., Meltzer, M. S., Phipps, T. and Gendelman, H. E. (1988). Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J Virol 62(8): 2578-2586.
- Pang, Y., Song, H. and Cheng, W. (2016). Using optical trap to measure the refractive index of a single animal virus in culture fluid with high precision. Biomed Opt Express 7(5): 1672-1689.
- Pillemer, E. A., Kooistra, D. A., Witte, O. N. and Weissman, I. L. (1986). Monoclonal antibody to the amino-terminal L sequence of murine leukemia virus glycosylated gag polyproteins demonstrates their unusual orientation in the cell membrane. J Virol 57(2): 413-421.
- Quek, C., Bellingham, S. A., Jung, C. H., Scicluna, B. J., Shambrook, M. C., Sharples, R. A., Cheng, L. and Hill, A. F. (2017). Defining the purity of exosomes required for diagnostic profiling of small RNA suitable for biomarker discovery. RNA Biol 14(2): 245-258.
- Raposo, G., Moore, M., Innes, D., Leijendekker, R., Leigh-Brown, A., Benaroch, P. and Geuze, H. (2002). Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3(10): 718-729.
- Raposo, G., Nijman, H. W., Stoorvogel, W., Liejendekker, R., Harding, C. V., Melief, C. J. and Geuze, H. J. (1996). B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183(3): 1161-1172.
- Rayaprolu, V., Ramsey, J., Wang, J.C. and Mukhopadhyay, S. (2018). Alphavirus purification using low-speed spin centrifugation. Bio-protocol 8(6): e2772.
- Renner, T. M., Belanger, K., Lam, C., Gerpe, M. C. R., McBane, J. E. and Langlois, M. A. (2018). Full-length glycosylated gag of murine leukemia virus can associate with the viral envelope as a Type I integral membrane protein. J Virol 92(6): e01530-17.
- Rosales Gerpe, M. C., Renner, T. M., Belanger, K., Lam, C., Aydin, H. and Langlois, M. A. (2015). N-linked glycosylation protects gammaretroviruses against deamination by APOBEC3 proteins. J Virol 89(4): 2342-2357.
- Sandrin, V. and Cosset, F. L. (2006). Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. J Biol Chem 281(1): 528-542.
- Sliva, K., Erlwein, O., Bittner, A. and Schnierle, B. S. (2004). Murine leukemia virus (MLV) replication monitored with fluorescent proteins. Virol J 1: 14.
- Tang, V. A., Renner, T. M., Fritzsche, A. K., Burger, D. and Langlois, M. A. (2017). Single-particle discrimination of retroviruses from extracellular vesicles by nanoscale flow cytometry. Sci Rep 7(1): 17769.
- Tang, V. A., Renner, T. M., Varette, O., Le Boeuf, F., Wang, J., Diallo, J. S., Bell, J. C. and Langlois, M. A. (2016). Single-particle characterization of oncolytic vaccinia virus by flow virometry. Vaccine 34(42): 5082-5089.
- Telesnitsky, A. and Wolin, S. L. (2016). The host RNAs in retroviral particles. Viruses 8(8): E235.
- van Niel, G., D'Angelo, G. and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19(4): 213-228.
- Wang, J. J., Horton, R., Varthakavi, V., Spearman, P. and Ratner, L. (1999). Formation and release of virus-like particles by HIV-1 matrix protein. AIDS 13(2): 281-283.
- Xu, R., Greening, D. W., Rai, A., Ji, H. and Simpson, R. J. (2015). Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87: 11-25.
- Yanez-Mo, M., Siljander, P. R., Andreu, Z., Zavec, A. B., Borras, F. E., Buzas, E. I., Buzas, K., Casal, E., Cappello, F., Carvalho, J., Colas, E., Cordeiro-da Silva, A., Fais, S., Falcon-Perez, J. M., Ghobrial, I. M., Giebel, B., Gimona, M., Graner, M., Gursel, I., Gursel, M., Heegaard, N. H., Hendrix, A., Kierulf, P., Kokubun, K., Kosanovic, M., Kralj-Iglic, V., Kramer-Albers, E. M., Laitinen, S., Lasser, C., Lener, T., Ligeti, E., Line, A., Lipps, G., Llorente, A., Lotvall, J., Mancek-Keber, M., Marcilla, A., Mittelbrunn, M., Nazarenko, I., Nolte-'t Hoen, E. N., Nyman, T. A., O'Driscoll, L., Olivan, M., Oliveira, C., Pallinger, E., Del Portillo, H. A., Reventos, J., Rigau, M., Rohde, E., Sammar, M., Sanchez-Madrid, F., Santarem, N., Schallmoser, K., Ostenfeld, M. S., Stoorvogel, W., Stukelj, R., Van der Grein, S. G., Vasconcelos, M. H., Wauben, M. H. and De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4: 27066.
- Yeager, M., Wilson-Kubalek, E. M., Weiner, S. G., Brown, P. O. and Rein, A. (1998). Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci U S A 95(13): 7299-7304.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Renner, T. M., Bélanger, K. and Langlois, M. (2018). Selective Isolation of Retroviruses from Extracellular Vesicles by Intact Virion Immunoprecipitation. Bio-protocol 8(17): e3005. DOI: 10.21769/BioProtoc.3005.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Microbiology > Microbial physiology > Membrane property
Molecular Biology > Protein > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












