- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Using Stable Isotopes in Bone Marrow Derived Macrophage to Analyze Metabolism
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.3003 Views: 8336
Reviewed by: Vivien Jane Coulson-ThomasMindy CallAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In-Gel Activity Assay of Mammalian Mitochondrial and Cytosolic Aconitases, Surrogate Markers of Compartment-Specific Oxidative Stress and Iron Status
Wing-Hang Tong and Tracey A. Rouault
Dec 5, 2024 2200 Views

Reliable and Sensitive Detection of Carbonylated Proteins by Oxime Blot
Filip Luka Mikulić [...] Mladen Merćep
Aug 5, 2025 1241 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2235 Views
Abstract
Using gas chromatography mass spectrometry (GC-MS) to analyze the citric acid cycle (CAC) and related intermediates (such as glutamate, glutamine, GABA, and aspartate) is an analytical approach to identify unexpected correlations between apparently related and unrelated pathways of energy metabolism. Intermediates can be as expressed as their absolute concentrations or relative ratios by using known amounts of added reference standards to the sample. GC-MS can also distinguish between heavy labeled molecules (2H- or 13C-labeled) and the naturally occurring most abundant molecules. Applications using tracers can also assess the turnover of specific metabolic pools under various physiological and pathological conditions as well as for pathway discovery.
The following protocol is a relatively simple method that is not only sensitive for small concentrations of metabolic intermediates but can also be used in vivo or in vitro to determine the integrity of various metabolic pathways, such as flux changes within specific metabolite pools. We used this protocol to determine the role of phosphoenolpyruvate carboxykinase 1 (Pck1) gene in mouse macrophage cells to determine the percent contribution from a precursor of 13C labeled glucose into specific CAC metabolite pools.
Background
With the development of altered gene expression in cells and mice, there is a need to understand how these deleted or over-expressed genes impact the regulation of metabolic pathways. In this protocol, we used stable isotopes to determine how the flux of glucose into the CAC altered the contribution of glucose into the pools of citrate, succinate and malate. The use of stable isotopes with targeted analysis of metabolism is just one benefit to using stable isotopes in cell culture.
The method described in this protocol for functional quantification of intracellular metabolites was done by growing bone marrow-derived macrophage cells (BMDM) in U-13C-glucose medium. The cells were extracted in organic solvent and the percent contribution of 13C glucose was calculated. The fractional amount of 13C label incorporated into each the CAC-related metabolite pools was also determined. The calculations were based on the ratio of 13C label of each intracellular metabolite versus the unlabeled metabolite; for absolute concentration analysis of cell samples, one would need to correct for reference to the intracellular volume of the extracted cells (Feldberg et al., 2009), as well as add non-interfering reference standards for quantifying of levels of CAC intermediates, as previously described (Ko et al., 2018). This protocol can also be used in isolated perfused livers or whole body metabolism studies (Yang et al., 2008a; Zhang et al., 2015).
Using a novel mouse model that had a deletion of phosphoenolpyruvate carboxykinase 1 (Pck1) in the myeloid cells (Pck1MC-KO), stable isotopes were used to determine the role of this gene in macrophages (Ko et al., 2018) with respect to glucose metabolism. The protocol explains the isolation and differentiation of BMDM. These cells were isolated, differentiated and incubated with U13C-glucose to analyze their metabolism. The BMDM cells were collected, and the fractional contribution of the precursor to the product was based on the mole percent enrichments (MPE) derived from the 13C label incorporation into the total pool of each of the metabolites (products). Mass isotopomer analysis enables the measurements of unlabeled analyte (M0) relative to the labeled analyte (M+1, 2, or 3, etc.), (Yang et al., 2008a; Kombu et al., 2011; Ko et al., 2018). The measured mass isotopomer distributions were calculated for each of the masses and expressed as mole percent enrichment (MPE) after correcting for natural isotope abundances.
Materials and Reagents
- FisherbrandTM sterile 100 mm x 15 mm polystyrene Petri dish (Fisher Scientific, Fisher ScientificTM, catalog number: FB0875713 )
- Costar® TC-Treated 6-well Plates (Corning, catalog number: CLS3506 )
- 15 ml conical tubes (SARSTEDT, catalog number: 62.554.502 )
- Sterile individually packaged 5 ml pipettes (SARSTEDT, catalog number: 86.1253.001 )
- Sterile 1 ml syringe with 26 G needle (Fisher Scientific, catalog number: 14-829-6A)
Manufacturer: BD, catalog number: 305537 . - 10 ml syringes (Thermo Fisher Scientific, catalog number: S7515-10 )
- BD Precisionglide® syringe needles, gauge 23, L3/4 in. (BD, catalog number: 305143 )
- BD Precisionglide® syringe needles, gauge 18, L 1 in. (BD, catalog number: 305195 )
- Cell strainer, 70 μm, sterile (Corning, catalog number: 352350 )
- 50 ml conical tube (SARSTEDT, catalog number: 62.547.254 )
- Disposable Borosilicate Glass tubes 16 mm x 125 mm (Globe Scientific, catalog number: 1515 )
- LysM-specific Pck1 knock-out mice (Pck1MC-KO mice)
Note: The mice were generated by crossing Pck1flox/flox mice with LysM-Cre+/− transgenic mice expressing Cre-recombinase under control of the LysM promoter. Pck1flox/flox mice were used as controls. All mice are in the C57Bl/6J background. All animals were housed in a temperature-controlled facility with a 12 h light/dark cycle in compliance with the Institutional Animal Care and Use Committee (IACUC) of Case Western Reserve University. - Lipopolysaccharides from Escherichia coli 026:B6 (Sigma-Aldrich, catalog number: L8274 )
- IL-4, Animal-component free, recombinant, expressed in E. coli (Sigma-Aldrich, catalog number: SRP3211 )
- Ethanol Solution 70%, Molecular Biology Grade (Fisher Scientific, Fisher BioagentsTM, catalog number: BP8201500 )
- FBS (fetal bovine serum) (Fisher Scientific, FisherbrandTM, catalog number: 03-600-511 )
- Dulbecco's modified Eagle Medium (DMEM) high glucose, pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11995065 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 10570063 )
- L-glutamine GlutaMAX (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Recombinant Murine Macrophage Colony-stimulating factor (PeproTech, catalog number: 315-02 )
- Glucose (Sigma-Aldrich, catalog number: G8270 )
- Lipopolysaccharide (LPS) from E. coli 02:B6 (Sigma-Aldrich, catalog number: L8274 )
- Interleukin-4 (IL-4) (PeproTech, catalog number: 214-14 )
- [13C]-succinate [Sodium bis (2-ethylhexyl) sulfo (succinate-13C4)] (98%) (Sigma-Aldrich, catalog number: 719269 )
- [13C]-malate (Sigma-Aldrich, catalog number: 750484 )
- [13C]-citrate (Sigma-Aldrich, catalog number: 492078 )
- Methanol HPLC (Sigma-Aldrich, catalog number: 1005706 )
- BSTFA + 10% TMCS-Regisil® (Regis Technologies, CAS: 25561-30-2; 75-77-4)
- N-Methyl-N-(t-butyldimethylsilyl) trifluoroacetamide (TBDMS) (Sigma-Aldrich, catalog number: 394882-25ML )
Note: If using TMCS as a derivative, see references Yang et al. (2008a); Kombu et al. (2011); Ko et al. (2018). - Acetic acid, Glacial (Certified ACS) Fisher Chemical (Fisher Scientific, catalog number: A38-212)
Manufacturer: Thermo Fisher Scientific, Thermo ScientificTM, catalog number: FLA38212 . - Methanol, OptimaTM LC/MS Grade, Fisher Chemical (Fisher Scientific, Thermo ScientificTM, catalog number: A456-1 )
- Water, OptimaTM LC/MS Grade, Fisher Chemical (Fisher Scientific, Thermo ScientificTM, catalog number: W64 )
- QuickStart TM Bradford Protein Assay Kit 1 (Bio-Rad Laboratories, catalog number: 5000201 )
- Generate macrophage differentiation media (MDM) (see Recipes)
- 5% acetic acid in methanol/water (1:1) extraction buffer (see Recipes)
- LPS stock solution (see Recipes)
- IL-4 stock solution (see Recipes)
Equipment
- Kelly Forceps, Box Lock, Straight Steel (Grainger, catalog number: 4WPD9 )
- Sterile cell scraper (Fisher Scientific, FisherbrandTM, catalog number: 08-100-241 )
- GC vial cap, 9 mm (Agilent Technologies, catalog number: 5182-0717 )
- GC vial, 2 ml (Agilent Technologies, catalog number: 5181-3375 )
- Thermo ScientificTM NalgeneTM Polypropylene Graduated Cylinders (Fisher Scientific, catalog number: 08-572D)
Manufacturer: Thermo Fisher Scientific, Thermo ScientificTM, catalog number: N36620100 . - Dumont forceps #5 (Fine Science Tools, catalog number: 11252-20 )
- Dumont forceps #55 (Fine Science Tools, catalog number: 11255-20 )
- Dumont forceps AA (Fine Science Tools, catalog number: 11210-20 )
- Tissue culture hood (Thermo Fisher Scientific, catalog number: 51022482 )
- Refrigerated tabletop centrifuge for 15-50 ml conical tubes (Eppendorf, model: 5430R )
- 37 °C, 5% CO2 water-jacketed incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: 3110 )
- P20 pipetman (Gilson, catalog number: F123600 )
- P200 pipetman (Gilson, catalog number: F123601 )
- P1000 pipetman (Gilson, catalog number: F123602 )
- Homogenizer for tissues (IKA, model: T25 digital ULTRA-TURRAX®, catalog number: 0003725001 )
- Dry Block Heater (VWR, catalog number: 75838-282 )
- Frigidaire 13.8 cu. Ft. Frost Free Upright Freezer (The Home Depot)
- Large Series, Single Door, Hinged Autoclave (Consolidated Sterilizer Systems, model: LR-36E )
- Basic Laboratory Hoods (Labconco, model: 2246500 )
- Inverted light microscope (Leica Microsystems, model: DM IL LED )
- GC-MS (Agilent Technologies, model: 5973-MSD ) equipped with an Agilent 6890 GC system
- DB-17MS capillary column, 30 m x 0.25 mm x 0.25 μm (Agilent Technologies)
Procedure
- Isolation and differentiation of Bone Marrow-Derived Macrophages and incubation with stable isotope
- Harvesting Marrow from Mouse Femur and Tibia
- Prepare macrophage differentiation media (MDM) (see Recipes). Warm up the media in a 37 °C water bath for 30 min before harvesting the marrow.
- Euthanize the mouse with CO2 asphyxiation and cervical dislocation and place the mouse in a beaker with 100% ethanol.
- Using sterile scissors in a tissue culture hood, clip the skin and remove the skin from the lower part of the mouse body.
- Remove the legs from the mouse body with scissors that have been placed in a beaker with 100% ethanol (Figure 1).
- Remove all remaining hair and skin. Remove muscle, and fat surrounding the bones. Place the bones in 100% ethanol for 5 min. Pour 8 ml of MDM into a sterile 100 mm x 15 mm polystyrene Petri dish. Transfer the bones from the 100% ethanol to the Petri dish with media.
- Separate at the knee joint the femoral bones from the tibia and fibula. Be careful not to break the bones.
- Cut off each end of the bone as shown in Figure 1. You should have a total of 3 bones for each mouse leg in one Petri dish with media.

Figure 1. Isolating bone marrow from mouse tibia, fibula and femur. The mouse was euthanized. A. Image illustrating where to cut and remove the leg. B. Schematic diagram of the positions to be cut at mouse leg for isolating bone marrow. Cut the legs at position 1 (red dashed lines). Next remove the fur, skin and muscle from the leg. Then cut the bones at the red dotted lines to isolate the bones in the order indicated (numbers 2, 3, 4 and 5). Note that each bone is cut medially and distally. - Fill 26 G needle/1 ml syringe with macrophage differentiation media (MDM) (see Recipes). Put the tip of the needle into the bone and flush the bone marrow from both ends of the bone (Figure 2). Do this for the femur, fibula and tibia. Repeat flushing the marrow out of the bones until the bones turn white.

Figure 2. Flushing the bones. A. Image showing the marrow compartment in the bone. B and C. The femur before (B) flushing and after (C) flushing (see Video 1).Video 1. Flushing of the bones to remove bone marrow - Collect the cells in MDM and pass them through a 70 μm cell strainer into a sterile conical tube. This removes debris that may contaminate your cells (see Video 2). Video 2. Removing debris from isolated cells
- Spin down cells at 1,950 x g for 5 min at 4 °C. Count the cells and resuspend the pellets with macrophage differentiation media (MDM) so that you can seed 5 x 106 cells and MDM into each well of the 6-well plate.
- Incubate plates for 7 days at 37 °C and 5% CO2. Change media every three days with freshly made media.
- Differentiation of macrophage to M1 or M2
When BMDM are almost confluent (at Day ~7), replace with new MDM. In order to polarize the macrophages, we used either lipopolysaccharide (LPS) or interleukin-4 (IL-4).- For classic M1 activation: Add lipopolysaccharide (LPS) at a dose of 100 ng/ml (10 μl of stock solution into 4 ml MDM) and incubate for 4 h.
- For classic M2 activation: Add interleukin-4 (IL-4) at a dose of 40 ng/ml (1.6 μl of stock solution into 4 ml of MDM) and incubate for 4 h.
- Metabolism studies using U-13C Glucose
- Replace the MDM in the differentiated macrophages (already treated with either LPS or IL-4) with medium that contains no glucose, 10% charcoal-stripped FBS, 100 ng/ml LPS, 5 mM U-13C glucose (99.98% enriched) and incubate overnight at 37 °C. We did this because the gene of being investigated, Pck1, is activated in the fasted state, so we removed the glucose and insulin.
- At the next day, collect the media and cells. To collect media, pipet off half of the media. Keep 1 ml of the media for measurement of isotope enrichment. To collect cells, scrape the wells to dislodge the cells. Pipet up the cells and remaining media and place into conical tubes. Spin down the cells at 1,950 x g for 5 min at 4 °C and resuspend the pellets in 50 μl of 1x PBS. Save 20 μl for measurement of protein concentration by Bradford assay. Freeze sample at -20 °C to lyse cells. The protein concentrations are used for normalization in the metabolite analysis. Cells are for analysis by GC-MS. In our case, we sent the cells to the Case Western Reserve University Mouse Metabolic Phenotyping Center (MMPC) for analysis by GC-MS.
- Bradford assay: Carry out the assay according to the manufacturer’s directions (Quick StartTM Bradford Protein Assay). We used 2 mg/ml BSA as a standard. Pipette 20 μl of the resuspended cells into a 1.5 ml Eppendorf tube and add 1 ml of 1x Dye reagent from the Quick StartTM Bradford Protein Assay. Mix the sample and incubate at room temperature for 5 min. Place the sample in a cuvette. Set the spectrophotometer to 595 nm. Zero the instrument with the blank sample (100 μl water and 1 ml of 1x Dye reagent). Measure the absorbance of the BSA standards and the unknown (macrophage cells). Determine the amount of protein in the sample from the standard curve (if 2 μl aliquot of sample yields a 595 nm value equivalent to 250 μg/ml of protein, then the cell sample would have a protein concentration of 2.5 mg/ml).
- For the study on Pck1MC-KO macrophages, the cells were sent to the Case Western Reserve University Mouse Metabolic Phenotyping Center (MMPC) for analysis by GC-MS. The following section displays an example for the study of metabolites in the LPS-activated macrophage. The purpose of the example study was to compare the metabolites in Pck1MC-KO (experimental) and Pck1fl/fl (control) macrophages after a 24 h treatment of LPS.
- Harvesting Marrow from Mouse Femur and Tibia
- Analysis of the citric acid cycle (CAC) and related intermediates (such as citrate, succinate and malate) using gas chromatography mass spectrometry (GC-MS)
- Cell sample preparation
Briefly, this approach utilizes the rapid reaction of silylating reagents with alcohols, acids and amines to form silyl derivatives (Yang et al., 2008a and 2008b; Kombu et al., 2011; Zhang et al., 2015). Commercial silylating reagents are available as combinations to accelerate the reaction, as well as to react with the hindered group. For example either a mixture of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) and trimethylchlorosilane (TMCS) to form a TMS derivative, or a mixture of N-Methyl-N-(t-butyldimethylsilyl)-trifluoroacetamide (MTBSTFA) and t-butyldimethylchlorosilane (TBDMS) to form TBDMS derivatives can be used (Yang et al., 2008a and 2008b; Kombu et al., 2011; Zhang et al., 2015).- For absolute quantification of analytes, as previously described (Ko et al., 2018): 50 μl of cells are spiked with selected internal reference standards such as ~5 nmole of each metabolite [13C6] citrate, [13C4] succinate, and/or [2H4]-3-hydroxyglutarate.
Note: Choice of internal reference standards are dependent on precursor tracer used such that there is no interference of the M+ labeling from the tracer versus the labeled standards added; its best to use non-endogenously produced reference standards, such as tricarballylic acid. - Homogenize the cells with 3-5 ml of 5% acetic acid in methanol/water (1:1) extraction buffer (chilled on ice) for 2 min on ice bath.
- Centrifuge the homogenate at 670 x g for 30 min at 4 °C. Decant the supernatant into a glass test tube and save on ice and process immediately
Note: May freeze the supernatant at -80 °C until assaying for GC-MS. - TMSC/TBDMS derivatization procedure: pipette 100-200 μl of the supernatant collected in Steps B1b and B1c into 16 mm x 125 mm disposable borosilicate glass tubes.
- Dry completely under nitrogen gas (Figure 3).

Figure 3. Nitrogen Dryer - React by adding 70 μl of either derivatizing TBDMS reagents (in a pyridine-based solvent) and heat on a heating block at 80 °C for 1 h (Figure 4).
Note: May react at room temperature overnight.
Figure 4. Heat block with tubes for derivatization - Transfer to GC insert and cap (Figure 5); follow GC parameters and the monitored SIM ions (m/z) for each analyte, as outlined (Yang et al., 2008a and 2008b; Kombu et al., 2011; Zhang et al., 2015).

Figure 5. Samples put into GC tubes
- For absolute quantification of analytes, as previously described (Ko et al., 2018): 50 μl of cells are spiked with selected internal reference standards such as ~5 nmole of each metabolite [13C6] citrate, [13C4] succinate, and/or [2H4]-3-hydroxyglutarate.
- GC-MS Analysis
- TMSC or TBDMS derivatives are analyzed using an Agilent 5973-MSD equipped with an Agilent 6890 GC system, and a DB-17MS capillary column (30 m x 0.25 mm x 0.25 μm; may use 60 m). The mass spectrometer is operated under electron impact mode (EI) or ammonia chemical ionization mode (CI) and selective ion monitoring (SIM) m/z for each analyte; When stable isotopes are applied, the SIM for M0-M+ m/z are monitored (Yang et al., 2008a and 2008b; Kombu et al., 2011; Zhang et al., 2015) (Figure 6).

Figure 6. GC-MS apparatus used in this protocol - After the GC-MS run, calculate the area under the curve for succinate, citrate and malate and the ratios to the internal standard (Table 1). After background correction, calculate the MPE% using the following formula:
MPE% = [(13C labeled M1 thru M + for each of the CAC intermediates) x (13C labeled + unlabeled) - 1] x 100 (Table 1)
Table 1. The calculation of mole percent enrichments from 13C-glucose into citrate, succinate and malate. M0, M1 to M3 were calculated based on the ratio of labeled intracellular metabolites to unlabeled and were corrected with reference to the intracellular volume of the extracted cells (Feldberg et al., 2009). The %MPE was calculated as %m2 = [(13C labeled M1 thru M + for each of the CAC intermediates) x (13C labeled + unlabeled) - 1] x 100.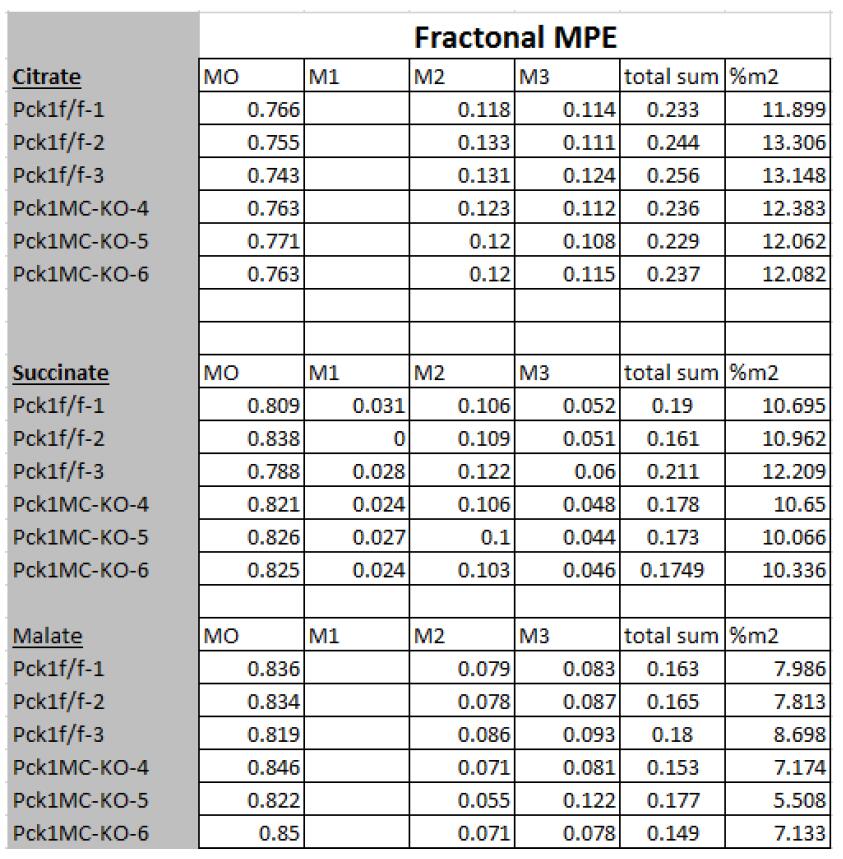
- TMSC or TBDMS derivatives are analyzed using an Agilent 5973-MSD equipped with an Agilent 6890 GC system, and a DB-17MS capillary column (30 m x 0.25 mm x 0.25 μm; may use 60 m). The mass spectrometer is operated under electron impact mode (EI) or ammonia chemical ionization mode (CI) and selective ion monitoring (SIM) m/z for each analyte; When stable isotopes are applied, the SIM for M0-M+ m/z are monitored (Yang et al., 2008a and 2008b; Kombu et al., 2011; Zhang et al., 2015) (Figure 6).
- Cell sample preparation
Data analysis
- For experimental design, the stable isotope studies are done in triplicate for each sample. The triplicates are considered an n = 1. All experiments are done for a total of n = 6 times.
- MPE of M1 to M3 was calculated based on the ratio of labeled intracellular metabolites to unlabeled and corrected with reference to the intracellular volume of the extracted cells (Feldberg et al., 2009). The %MPE was calculated as MPE% = [(13C labeled M1 thru M+ for each of the CAC intermediates) x (13C labeled + unlabeled)] x 100.
- For statistical analysis, we used GraphPad Prism. To compare two data sets (Pck1flox/flox and Pck1MC-KO mice), we performed a Student's t-test (Figure 7).
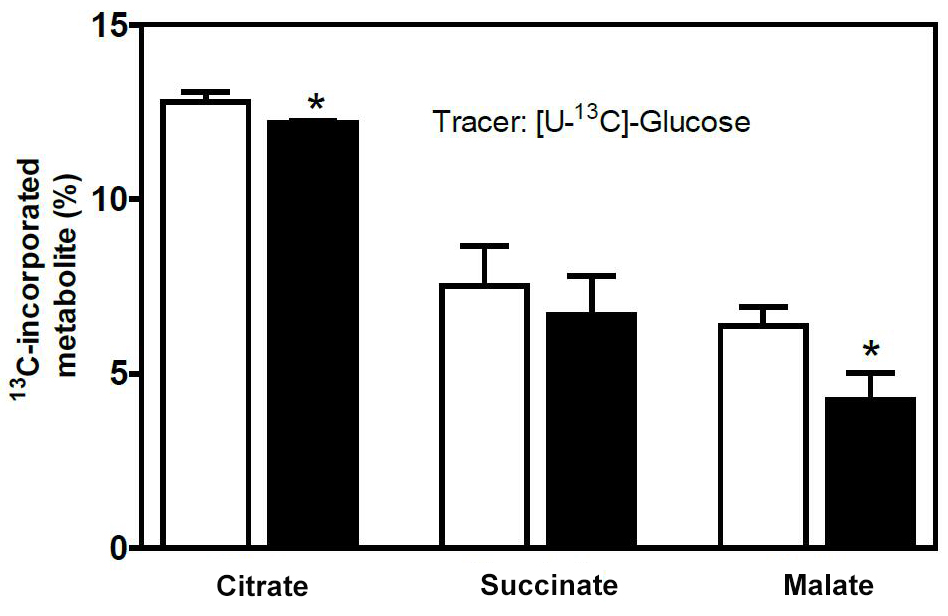
Figure 7. Contribution of glucose to the central metabolism of LPS-activated macrophages. BMDMs isolated from Pck1flox/flox and Pck1MC-KO mice were incubated with [U-13C] glucose in the presence of LPS for 16 h, and GC-MS was performed for the calculation of the contribution of 13C to M2-CAC. Values represent the means ± SEM for n = 6 per group. *P < 0.05 compared to Pck1flox/flox group.
Notes
- Internal standards are necessary due to variation in each run. For this experiment, the samples (n = 6 per group) were all run on the GC-MS at the same time to reduce variation.
- Isolation of BMDM was done for each animal one at a time. For example, one Pck1flox/flox mouse was euthanized, legs dissected and bone marrow was flushed from the femur tibia and fibula. The cells were kept cold on ice. Then the next animal was euthanized and so on. The cells were kept on ice until all of the bone marrow was collected.
- Media was prewarmed before plating of BMDM.
- To study the initiation of signaling pathways (e.g., TLR4 signaling in M1 activation or IL-4R signaling in M2 activation) that require rapid turnover, cells are suggested to be treated with higher dose of stimulus (e.g., 100 ng/ml LPS or 20 ng/ml IL-4) within 30 min; to determine the metabolic pathways (e.g., citric acid cycle and related intermediates) using isotopomers, 16-24 h incubation with lower dose of stimulus (e.g., 20 ng/ml LPS or 10 ng/ml IL-4) is suggested.
- Macrophages undergo morphological changes during their activation. Compared to the non-stimulated macrophage with rounded shapes, LPS-treated macrophages display pancake-like shape within 24 h of stimulation. On the other hand, IL-4-treated cells promote cell elongation.
- The cells isolated from the bone marrow will adhere to the tissue culture plates within 24 h-48 h. The MDM contains M-CSF that will allow for myeloid cells to be differentiated into macrophages. If the cells continue to float in the media there may be contamination of the cells. If this occurs, you will need fresh media and re-isolate the bone marrow cells.
Recipes
- Generate macrophage differentiation media (MDM)
Combine 500 ml of sterile Dulbecco’s modified Eagle Medium (DMEM) with:
55 ml of 10% fetal bovine serum (FBS) to the DMEM
5 ml of 1x glutamine to DMEM
5 ml of 1x Penicillin-Streptomycin (10,000 U/ml Pen, 10 mg/ml Strep)
5 μg of macrophage colony-stimulating factor (M-CSF) to 500 ml of DMEM - 5% acetic acid in methanol/water (1:1) extraction buffer (chilled on ice)
Make methanol/water by adding 50 ml of methanol to 50 ml of dH2O to make 100 ml
In a separate graduated cylinder, combine 5 ml of acetic acid with 95 ml of the methanol/water made above - LPS stock solutions
Make up to 40 µg/µl in sterile water and aliquot
Store at -80 °C - IL-4 stock solution
Make up to 100 µg/µl in sterile water and aliquot
Store at -80 °C
Acknowledgments
The Case MMPC was supported by NIH grant U24 DK76174. The complete study on Pck1MC-KO mice has been published (Ko et al., 2018).
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
Ethics
All animal experiments were approved by Case Western Reserve University Institutional Animal Care and Use Committee (IACUC).
References
- Feldberg, L., Venger, I., Malitsky, S., Rogachev, I. and Aharoni, A. (2009). Dual labeling of metabolites for metabolome analysis (DLEMMA): A new approach for the identification and relative quantification of metabolites by means of dual isotope labeling and liquid chromatography-mass spectrometry. Anal Chem 81(22): 9257-9266.
- Ko, C. W., Counihan, D., Wu, J., Hatzoglou, M., Puchowicz, M. A. and Croniger, C. M. (2018). Macrophages with a deletion of the phosphoenolpyruvate carboxykinase 1 (Pck1) gene have a more proinflammatory phenotype. J Biol Chem 293(9): 3399-3409.
- Kombu, R. S., Brunengraber. H. and Puchowicz, M. A. (2011). Analysis of the citric acid cycle intermediates using gas chromatography-mass spectrometry. In: Metz, T. O. (Ed.) Metabolic Profiling: Methods and Protocols. Humana Press 147-157.
- Yang, L., Kasumov, T., Kombu, R. S., Zhu, S. H., Cendrowski, A. V., David, F., Anderson, V. E., Kelleher, J. K. and Brunengraber, H. (2008a). Metabolomic and mass isotopomer analysis of liver gluconeogenesis and citric acid cycle: II. Heterogeneity of metabolite labeling pattern. J Biol Chem 283(32): 21988-21996.
- Yang, L., Kombu, R. S., Kasumov, T., Zhu, S. H., Cendrowski, A. V., David, F., Anderson, V. E., Kelleher, J. K. and Brunengraber, H. (2008b). Metabolomic and mass isotopomer analysis of liver gluconeogenesis and citric acid cycle. I. Interrelation between gluconeogenesis and cataplerosis; formation of methoxamates from aminooxyacetate and ketoacids. J Biol Chem 283(32): 21978-21987.
- Zhang, Y., Zhang, S., Marin-Valencia, I. and Puchowicz, M. A. (2015). Decreased carbon shunting from glucose toward oxidative metabolism in diet-induced ketotic rat brain. J Neurochem 132(3): 301-312.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ko, C. W., Counihan, D., DeSantis, D., Sedor-Schiffhauer, Z., Puchowicz, M. and Croniger, C. M. (2018). Using Stable Isotopes in Bone Marrow Derived Macrophage to Analyze Metabolism. Bio-protocol 8(17): e3003. DOI: 10.21769/BioProtoc.3003.
- Ko, C. W., Counihan, D., Wu, J., Hatzoglou, M., Puchowicz, M. A. and Croniger, C. M. (2018). Macrophages with a deletion of the phosphoenolpyruvate carboxykinase 1 (Pck1) gene have a more proinflammatory phenotype. J Biol Chem 293(9): 3399-3409.
Category
Immunology > Immune cell isolation > Macrophage
Cell Biology > Cell metabolism > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










