- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enzymatic Assays and Enzyme Histochemistry of Tuta absoluta Feeding on Tomato Leaves
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.2993 Views: 7614
Reviewed by: Samik BhattacharyaSaptashati BiswasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1466 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3197 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1812 Views
Abstract
Enzymes play a key role in insect-plant relationships. For a better understanding of these interactions, we analyzed Tuta absoluta digestive enzymes. Here, we describe a detailed protocol for the detection of trypsin and papain-like enzymes in Tuta absoluta larvae by enzyme histochemistry. This assay uses frozen and unfixed samples to avoid the loss of enzymatic activity. We also describe a protocol for the quantification of trypsin and papain-like enzymes in the larvae of Tuta absoluta at different developmental instars.
Keywords: Tuta absolutaBackground
Plants and insects have coexisted for million years and evolved a set of interactions which affect both organisms at different levels. Insects manage to develop different physiological and morphological adaptations to overcome plants defense mechanisms. Therefore, a better understanding of their vital functions would facilitate their targeted control. Different physiological functions in insects rely on enzymes: digestive, respiratory, circulatory, muscular, nervous, reproductive and endocrine. Several enzymes participate in the digestion. Proteases such as trypsin, chymotrypsin, pepsin or carboxypeptidases are responsible for protein digestion, which is a source of amino acids for the insect. Thus, digestive protease inhibitors have been successfully used to improve plant resistance to insects (Smigocki et al., 2013; Quilis et al., 2014; Hamza et al., 2018). In order to design such approaches, it is important to identify the target insect digestive enzymes. Enzyme histochemistry is a useful method for the localization of active enzymes in tissue sections of an organism. However, few protocols have been described for insects. One of the main issues of this method is the sensibility of the enzymes to fixatives. In a previous study, Erban and Hubert (2011) visualized the digestive enzymes in the body of the acarid mite Lepidoglyphus destructor after feeding them with chromogenic and fluorescent substrates taking advantage of their transparent body. This approach is not suitable for larvae of bigger insects such as Tuta absoluta. These larvae do not have transparent body allowing the visualization of the fluorescence and are reared with fresh tomato leaves instead of artificial diet and thus cannot be supplemented with enzymatic substrates. In our work, we designed a protocol for the detection of serine proteases and papain-like proteases in cryosections of larvae of Tuta absoluta without tissue fixation. We also quantified these enzymes in larvae at different instars using chromogenic substrates.
Materials and Reagents
- 1.5 ml microcentrifuge tubes
- Petri dishes (Thermo Fisher Scientific, SterilinTM, catalog number: 122TS1 )
- 1.5 ml pestles (Sigma-Aldrich, catalog number: Z359947-100EA )
- Tuta absoluta eggs
Note: The eggs were obtained from the Tuta absoluta colony reared in the IVIA (Instituto Valenciano de Investigaciones Agrarias). The colony was started with adults caught from tomato fields near Castellon (Spain). No commercial insects/eggs are available. - Tomato leaves (cv. Micro-Tom)
- Liquid nitrogen (commercial grade)
- European Bacteriological Agar (Conda, catalog number: 1800 )
- Acetone (Merck, catalog number: 1000141000 , CAS: 67-64-1)
- Bovine serum albumin (BSA Fraction V) (Roche, catalog number: 10 735 078 001 )
- Bradford reagent (Bio-Rad Laboratories, catalog number: 5000006 )
- Trichloroacetic acid (AppliChem, Panreac, catalog number: 131067 , CAS: 76-03-9)
- Nα-Benzoyl-L-arginine 4-nitroanilide hydrochloride (BApNA) (Sigma-Aldrich, catalog number: B3133 , CAS: 21653-40-7)
- pGlu-Phe-Leu p-nitroanilide (Sigma-Aldrich, catalog number: P3169 , CAS: 85901-57-1)
- Bovine trypsin (Sigma-Aldrich, catalog number: T1426 , CAS: 9002-07-7)
- Papain (Sigma-Aldrich, catalog number: P4762 , CAS: 9001-73-4)
- Nα-Benzoyl-L-arginine-7-amido-4-methylcoumarin hydrochloride (BAAMC) (Santa Cruz Biotechnology, catalog number: sc-301455 , CAS: 83701-04-6)
- NEG-50 gel (Thermo Fisher Scientific, catalog number: 6502 )
- Pills blister (capacity: approximately 400 µl) (Enantyum 25 mg, Menarini)
- Polyvinyl alcohol (Sigma-Aldrich, catalog number: P1763 , CAS: 9002-89-5)
- L-Cysteine (Sigma-Aldrich, catalog number: 168149 , CAS: 52-90-4)
- Ascorbic acid (Duchefa Biochemie, catalog number: A0602 , CAS: 50-81-7)
- Tris (Duchefa Biochemie, catalog number: T1501.1000 , CAS: 77-86-1)
- Sucrose (AppliChem, catalog number: 131621.1211 , CAS: 57-80-1)
- Polyvinylpyrrolidone (PVP) (Sigma-Aldrich, catalog number: PVP10 , CAS: 9003-39-8)
- Sodium phosphate (AppliChem, Panreac, catalog number: 122018.1210 , CAS: 7601-54-9)
- Calcium chloride 2 hydrate (AppliChem, Panreac, catalog number: 131232.1210 , CAS: 10035-04-8)
- Protein extraction buffer (see Recipes)
- Trypsin assay buffer (see Recipes)
- Papain assay buffer (see Recipes)
- Histochemistry wash solution 1 (see Recipes)
- Histochemistry wash solution 2 (see Recipes)
- Histochemistry substrate solution (see Recipes)
Equipment
- Pipettes (Gilson, 2-20,20-200 and 100-1,000 µl)
- Growth chamber (SANYO, model: MLR 350 )
- Binocular stereomicroscope (Olympus, model: SZ-St )
- Brush (Staedtler permanent, model: 989 4 BK2 )
- Stainless steel tweezers
- Spectrophotometer (Eppendorf, model: 6131 )
- Cryostat (Thermo Fisher Scientific, MICROM, model: HM520 )
- Refrigerated centrifuge (Eppendorf, model: 5417R )
- Fluorescence microscope (Leica, model: DM5000 B )
- Poly-lysine coated slides (Thermo Fisher Scientific, catalog number: J2800AMNZ )
- Camera (Nikon, model: D3200 )
Procedure
- Tuta absoluta larvae Cryo-sectioning (Video 1)Video 1. Sample preparation and cryostat sectioning of Tuta absoluta larvae
- Collect Tuta absoluta larvae into 1.5 ml tubes and freeze them in liquid nitrogen.
- Pour a small volume of NEG-50 cryo-protector gel in a mold. Use a pills blister as a mold.
- Deposit the frozen larva in the mold and cover it with cryo-protective gel.
Note: Make sure there are no air bubbles in the gel. - Freeze the mold with the gel-included larva in liquid nitrogen until all the air bubbles are removed.
- Remove the frozen sample from the mold.
- Cover the surface of a pre-chilled cryostat specimen disc with NEG-50 gel.
- Deposit the frozen sample on the disc and place it in the cryostat chamber to freeze.
- Once all the gel is frozen, fix the specimen disc in the orientable specimen head.
- Set the section thickness to 16 μm and proceed to sectioning.
- Once it reaches the region of interest of the insect and you are able to see the digestive system of the larva, recover the section on a poly-lysine coated slide.
- Enzyme histochemistry
- Incubate the slide with 1 ml of the Histochemistry wash solution 1 for 2 min at room temperature to avoid macromolecules diffusion.
- Deposit 50 μl of the Histochemistry substrate solution on the cryo-section and incubate at 37 °C for 15 min in the dark.
- Wash the slide 5 times each with 1 ml of the Histochemistry wash solution 2. Keep the slides at -20 °C until you observe them under a microscope. You can keep the slides for several days at -20 °C.
- Total protein extraction
- Deposit one egg of Tuta absoluta on a fresh tomato leaf and incubate in a Petri dish with 2% agar (Figure 1).

Figure 1. Larva of Tuta absoluta in the third instar (L3) developing in a tomato leaf. The leafminer larva feeds and develops inside tomato leaves protected from predators and insecticides (Camera: Nikon D3200). - Follow the larval development daily under a binocular stereomicroscope.
Larvae of the first instar (L1) have a creamy yellowish color with a dark head and measure about 1.6 mm. Larvae of the second instar (L2) are about 2.8 mm long. In the third instar (L3), larvae become greener and increase their size reaching 4.7 mm. In the fourth larval instar (L4), it acquires a dorsal red colored band. In this last larval instar, it can reach 9 mm (Figure 2) (EPPO, 2011).
Figure 2. Larvae of Tuta absoluta at different development instars. L1: first instar; L2: second instar; L3: third instar; L4: fourth instar. During its development, the larva becomes bigger and acquires a green color. - One day after each molting, recover the larva and sacrifice it by freezing in liquid nitrogen.
- Pool about 40 mg of larvae of each instar in 1.5 ml microcentrifuge tubes.
- Grind larvae in liquid nitrogen.
- Homogenize the obtained powder in 200 μl of ice cold Protein extraction buffer.
- Centrifuge at 16,200 x g for 15 min at 4 °C.
- Recover the supernatant, mix it with 400 μl of 90% ice cold acetone and incubate at -20 °C for 2 h.
- Centrifuge at 16,200 x g for 10 min at 4 °C and discard the supernatant.
- Wash the pellet twice with 90% ice cold acetone and resuspend it in 100 μl of 0.5 M Tris buffer pH 8.
- Measure protein concentration in the crude extract by the Bradford method (Bradford, 1976).
- Deposit one egg of Tuta absoluta on a fresh tomato leaf and incubate in a Petri dish with 2% agar (Figure 1).
- Preparation of trypsin and papain standard curve
- Prepare trypsin and papain standards: 0.5 μg/μl, 1.0 μg/μl, 1.5 μg/μl, 2.0 μg/μl, and 2.5 μg/μl.
- Add 50 μg of the substrate to the standards (BApNA to trypsin and PFLNA to papain), and make up the volume to 100 μl with trypsin or papain assay buffer, respectively.
- Incubate at 37 °C for 30 min.
- Measure absorbance at 405 nm with a spectrophotometer.
- Quantification of trypsin and papain content
- Mix 5 μg of protein extract, 50 μg of the substrate and up to 100 μl of trypsin/papain assay buffer.
- Incubate at 37 °C for 30 min.
- Centrifuge the tubes at 16,200 x g for 10 min at 4 °C and recover the supernatant.
- Measure absorbance at 405 nm with a spectrophotometer.
Data analysis
- Quantification of trypsin and papain content
The concentration of trypsin and papain in the samples are calculated based on the standard curve (Figure 3).
The results are expressed as the percentage of trypsin or papain-like proteins from the total proteins of the crude extract (Figure 4).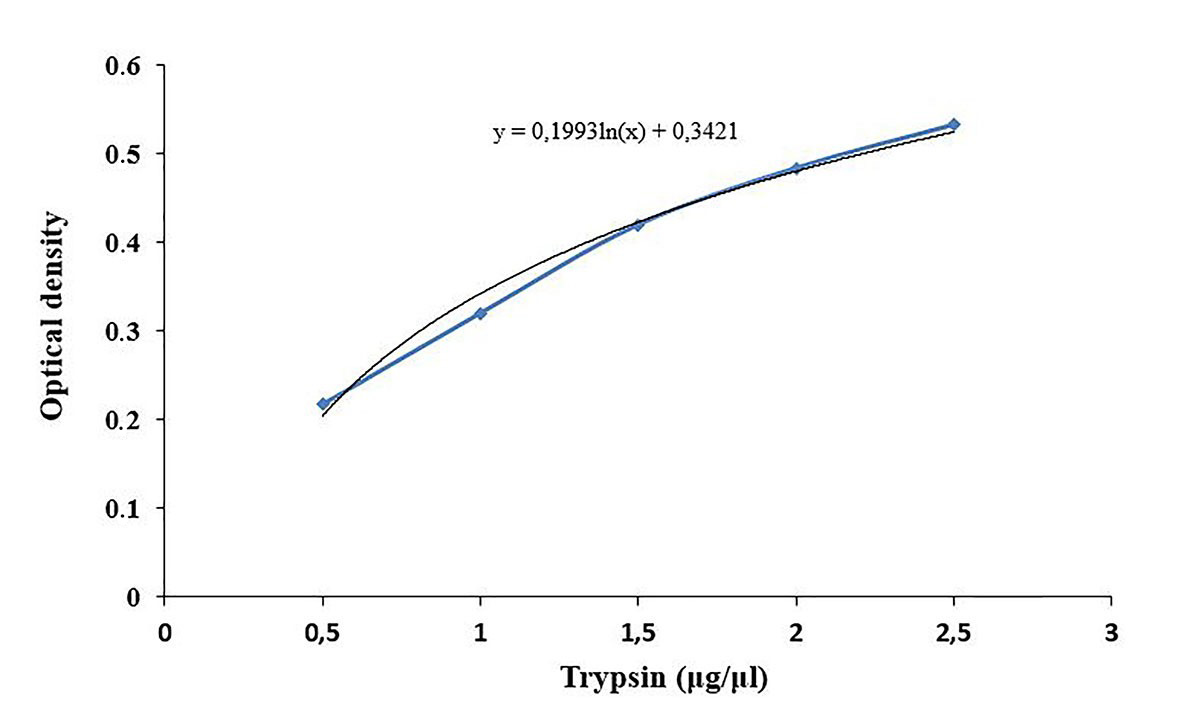
Figure 3. Standard curve for trypsin content determination. Commercial trypsin is used at different concentrations to obtain the standard curve.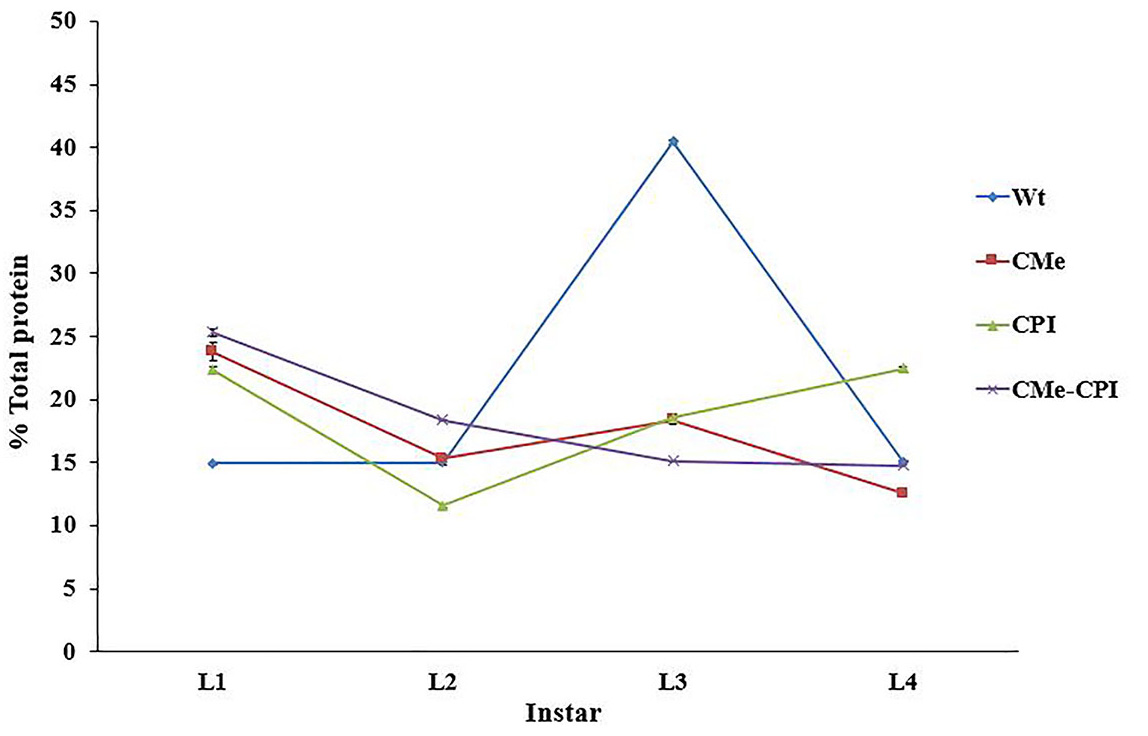
Figure 4. Trypsin content in Tuta absoluta larvae fed with different transgenic tomato leaves. Trypsin content is expressed as the percentage of trypsin-like proteins from the total proteins of the crude extract. - Enzyme histochemistry
When the substrate (Nα-Benzoyl-L-arginine-7-amido-4-methylcoumarin hydrochloride: BAAMC) binds to trypsin or papain-like proteases, it emits blue fluorescence. The fluorescence is observed with a Leica DM5000 microscope under ultraviolet light (Figure 5) (Objective: 5x/0.15 HC PL FLUOTAR, Exposure time: 1.8 sec, Gain: 1.5x).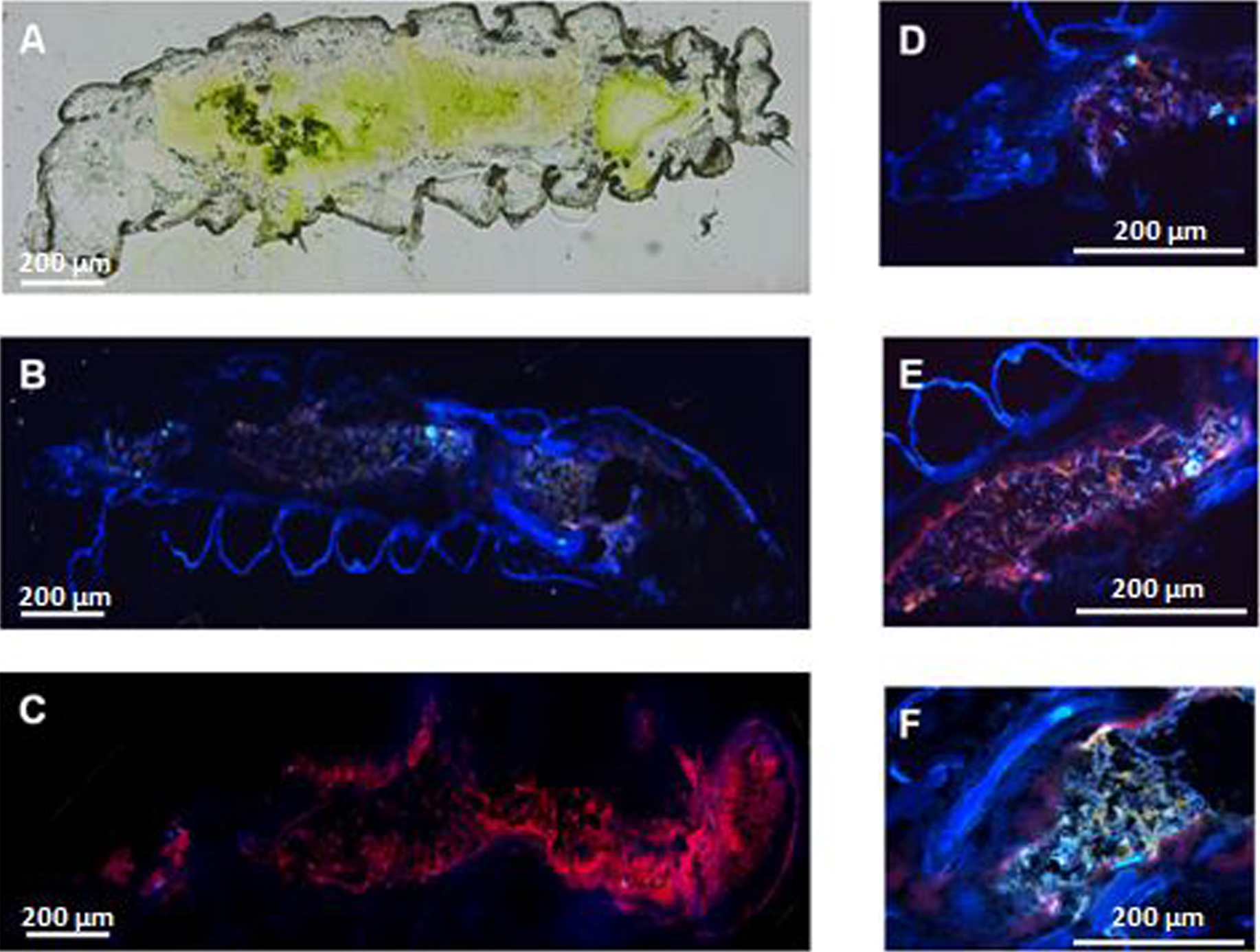
Figure 5. Enzyme histochemistry: detection of trypsin and papain-like enzymes in larvae of Tuta absoluta. A. Cryosection of Tuta absoluta L3 larva. B. Larval section incubated with BAAMC fluorescent substrate. C. Negative control: larval section without BAAMC substrate. D. Detail of the larval foregut. E. Detail of the larval midgut. F. Detail of the larval hindgut and Malpighi tubules.
Recipes
- Protein extraction buffer
0.1 M Tris pH 7
0.1% Ascorbic acid
0.1% L-Cysteine
0.5 M Sucrose
10 mg/ml Polyvinylpyrrolidone (PVP) - Trypsin assay buffer
Sodium phosphate buffer 67 mM, pH 7.6
20 mM CaCl2 - Papain assay buffer
Sodium phosphate buffer 67 mM, pH 7.6
5 mM L-Cysteine - Histochemistry wash solution 1
10% Polyvinyl alcohol
Sodium phosphate buffer 67 mM, pH 7.6 - Histochemistry wash solution 2
5% Polyvinyl alcohol
Sodium phosphate buffer 67 mM, pH 7.6 - Histochemistry substrate solution
10% Polyvinyl alcohol
0.5 μl BAAMC (20 mg/ml)
2 mM CaCl2
Sodium phosphate buffer 67 mM, pH 7.6
Acknowledgments
Rim Hamza acknowledges fellowships from the Tunisian Ministry for Higher Education and Scientific Research and from the Erasmus Mundus EMMAG program of the European Union. This work was partly supported by grants BIO2013-40747-R and AGL2014-55616-C3 from the Spanish Ministry of Economy and Competitiveness (MINECO). We wish to thank Drs. Alberto Urbaneja and Meritxell Pérez-Hedo (Instituto Valenciano de Investigaciones Agrarias, Centro de Protección Vegetal y Biotecnología, Unidad Asociada de Entomología, UJI-IVIA) for providing Tuta absoluta larvae and to Marisol Gascón (IBMCP) for technical assistance in sample processing. This protocol has been described in the publication: Hamza et al. (2018).
Competing interests
The authors declare that there are no conflicts of interest related with this work.
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- EPPO. (2011). First report of Tuta absoluta in Iraq. EPPO Reporting Service 4(5).
- Erban, T. and Hubert, J. (2011). Visualization of protein digestion in the midgut of the acarid mite Lepidoglyphus destructor. Arch Insect Biochem Physiol 78(2): 74-86.
- Hamza, R., Perez-Hedo, M., Urbaneja, A., Rambla, J. L., Granell, A., Gaddour, K., Beltran, J. P. and Cañas, L. A. (2018). Expression of two barley proteinase inhibitors in tomato promotes endogenous defensive response and enhances resistance to Tuta absoluta. BMC Plant Biol 18(1): 24.
- Quilis, J., Lopez-Garcia, B., Meynard, D., Guiderdoni, E. and San Segundo, B. (2014). Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant Biotechnol J 12(3): 367-377.
- Smigocki, A. C., Ivic-Haymes, S., Li, H. and Savic, J. (2013). Pest protection conferred by a Beta vulgaris serine proteinase inhibitor gene. PLoS One 8(2): e57303.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hamza, R., Beltrán, J. P. and Cañas, L. A. (2018). Enzymatic Assays and Enzyme Histochemistry of Tuta absoluta Feeding on Tomato Leaves. Bio-protocol 8(17): e2993. DOI: 10.21769/BioProtoc.2993.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link













