- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of the Effect of Sphingomyelinase on Rubella Virus Infectivity in Two Cell Lines
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.2992 Views: 6086
Reviewed by: Vamseedhar RayaproluVaibhav B ShahSaumik Basu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Production and Purification of Cell Culture–generated Hepatitis B Virus by Transient Transfection and Density Gradient
Asako Murayama [...] Takanobu Kato
Jul 20, 2023 1899 Views

An Improved Focus-Forming Assay for Determination of the Dengue Virus Titer
Maharah Binte Abdul Mahid [...] Kitti Wing Ki Chan
Oct 20, 2024 2396 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2457 Views
Abstract
Rubella is a mildly contagious disease characterized by low-grade fever and a morbilliform rash caused by the rubella virus (RuV). Viruses often use cellular phospholipids for infection. We studied the roles of cellular sphingomyelin in RuV infection. Treatment of cells with sphingomyelinase (SMase) inhibited RuV infection in rabbit kidney-derived RK13 cells and African green monkey (Cercopithecus aethiops) kidney-derived Vero cells. Our data further demonstrated that RuV used cellular sphingomyelin and cholesterol for its binding to cells and membrane fusion at the step of virus entry. Detailed protocols of our assays, which assess the effects of SMase treatment on RuV infectivity in RK13 and Vero cells, are described.
Keywords: Rubella virusBackground
Rubella virus (RuV) is a positive-strand RNA virus and belongs to the genus Rubivirus in the family Togaviridae. The family has two genera, Rubivirus and Alphavirus. RuV is the sole member of genus Rubivirus, whereas many viruses, such as Semliki forest virus (SFV) and Sindbis virus (SINV), are classified in the genus alphavirus. RuV is the causative agent of rubella and congenital rubella syndrome (CRS). Rubella is characterized by low-grade fever, a morbilliform rash, and lymphadenopathy. It is ordinarily a mild disease. However, CRS is a serious disease. CRS causes multiple organ defects in neonates born from mothers who suffered from rubella during the early phase of their pregnancy. Cataracts, sensorineural hearing loss and cardiovascular defects are common in CRS.
Previous studies suggested that cellular membrane lipids act as binding or entry factors for RuV infection (Mastromarino et al., 1989 and 1990), but the detailed mechanism has not yet been elucidated. Myelin oligodendrocyte glycoprotein (MOG) has been identified as a cellular receptor for RuV (Cong et al., 2011). However, the pathology of rubella cannot be solely explained by the usage of MOG because MOG is mainly expressed in the central nervous system and is barely expressed in other organs. Trinh et al. (2018) recently reported that RuV infects HaCat keratinocyte cells that do not express MOG on their surface.
Our recent study (Otsuki et al., 2018) demonstrated that RuV binds directly to sphingomyelin (SM) and cholesterol (SM/Chol)-enriched membranes in a Ca2+-dependent manner. Furthermore, the study showed that the binding is essential for membrane fusion at the early stage of rubella infection. In the current protocol, we provide a detailed method to examine whether the SM of host cells is essential for RuV infection in adherent cell lines. This protocol will be also useful to evaluate the use of host cellular SM in the infection of other viruses.
Materials and Reagents
- Materials
- A 6-well cell culture plate (Corning, catalog number: 3506 )
- A 12-well cell culture plate (Corning, catalog number: 3512 )
- A 24-well cell culture plate (Corning, catalog number: 3524 )
- Pipette tips (Neptune Scientific, catalog numbers: BT20 , BT200 and BT1250 )
- A 1.5 ml tube with lid (INA•OPTIKA, catalog number: ST-0150F )
- Cells
- RK13 cells: these cells were a gift from the Kitasato Institute
Note: These cells are maintained in Eagle’s minimum essential medium (MEM) (see Recipes) containing 8% bovine serum (BS). - Vero cells: our pre-existing stocks were originally obtained from American Type Culture Collection and have been maintained for more than 20 years in our laboratory
Note: These cells are maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% fetal bovine serum (FBS), 100 unit/ml Penicillin, and 100 μg/ml Streptomycin.
- RK13 cells: these cells were a gift from the Kitasato Institute
- Viruses
- Rubella virus (RuV) TO-336 vaccine strain (Takeda Pharmaceutical) (Otsuki et al., 2011)
- RuV RVi/Hiroshima.JPN/01.03 wild type strain (Otsuki et al., 2011; Sakata et al., 2014)
- The recombinant RuV RVi/Hiroshima.JPN/01.03 strain expressing the green fluorescent AG1 protein (rHS/p150-AG1) (reported previously in Sakata et al., 2014)
- The recombinant measles virus (MeV) and human metapneumovirus (HMPV) expressing enhanced green fluorescent protein (EGFP) (MeV-IC323/Ed-H-EGFP and HMPV-rJPS02-76EGFP, respectively) (reported previously in Seki et al., 2006 and Shirogane et al., 2008)
Notes:- The TO-336 vaccine strain and RVi/Hiroshima.JPN/01.03 wild type strain were propagated in RK13 cells. The recombinant rHS/p150-AG1 was propagated in Vero cells. MeV and HMPV were propagated in Vero/hSLAM cells (Ono et al., 2001) and Vero/TMPRSS2 cells (Shirogane et al., 2008), respectively.
- Viruses were diluted with MEM before use.
- Virus-like particles of RuV (RuV-VLP)
RuV-VLP, whose genome encodes Renilla luciferase and AG1 reporter genes, were produced and concentrated as described previously (Sakata et al., 2014), with some modifications (Otsuki et al., 2018). - Reagents
- Eagle’s MEM “Nissui” 1 (NISSUI PHARMACEUTICAL, catalog number: 05900 )
- Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, catalog number: D5796 )
- Dulbecco’s phosphate buffered saline (DPBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190144 )
- Fetal bovine serum (Biowest, catalog number: S1780-500 )
- Bovine serum (Thermo Fisher Scientific, GibcoTM, catalog number: 16170078 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- L-Glutamine (200 mM) (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Agarose ME (IWAI CHEMICALS, catalog number: 50013R )
- 7.5% sodium bicarbonate (Thermo Fisher Scientific, GibcoTM, catalog number: 25080094 )
- Sphingomyelinase from Bacillus cereus (Sigma-Aldrich, catalog number: S9396 )
- Neutral red solution (0.33%) (Sigma-Aldrich, catalog number: N2889 )
- Renilla Luciferase Assay system (Promega, catalog number: E2810 )
Note: Lysis buffer and Renilla Luciferase Assay Reagent are included in this kit. - MEM (see Recipes)
- 2x MEM (see Recipes)
- MEM containing 2% BS (or FBS) and 0.5% (or 0.4%) agarose (see Recipes)
- MEM containing 0.01% neutral red and 0.5% agarose (see Recipes)
Equipment
- Micropipettes (Gilson, models: P10, P200, P1000, catalog numbers: F144802 , F123601 , F123602 )
- Luminometer (Promega, model: GloMax® 20/20 )
- Autoclave (TOMY SEIKO, model: ES-215 )
- Biosafety cabinet (Air Tech)
- Cell culture incubator (at 35 °C and 37 °C with 5% CO2) (Thermo Fisher Scientific, Forma, model: 310 )
- Fluorescence microscope (ZEISS, model: Axio Observer D1 )
- Cell culture microscope (Olympus, model: CKX53 )
Procedure
- Analysis of the effect of SMase treatment on viral infectivity in RK13 cells (Figure 1A).
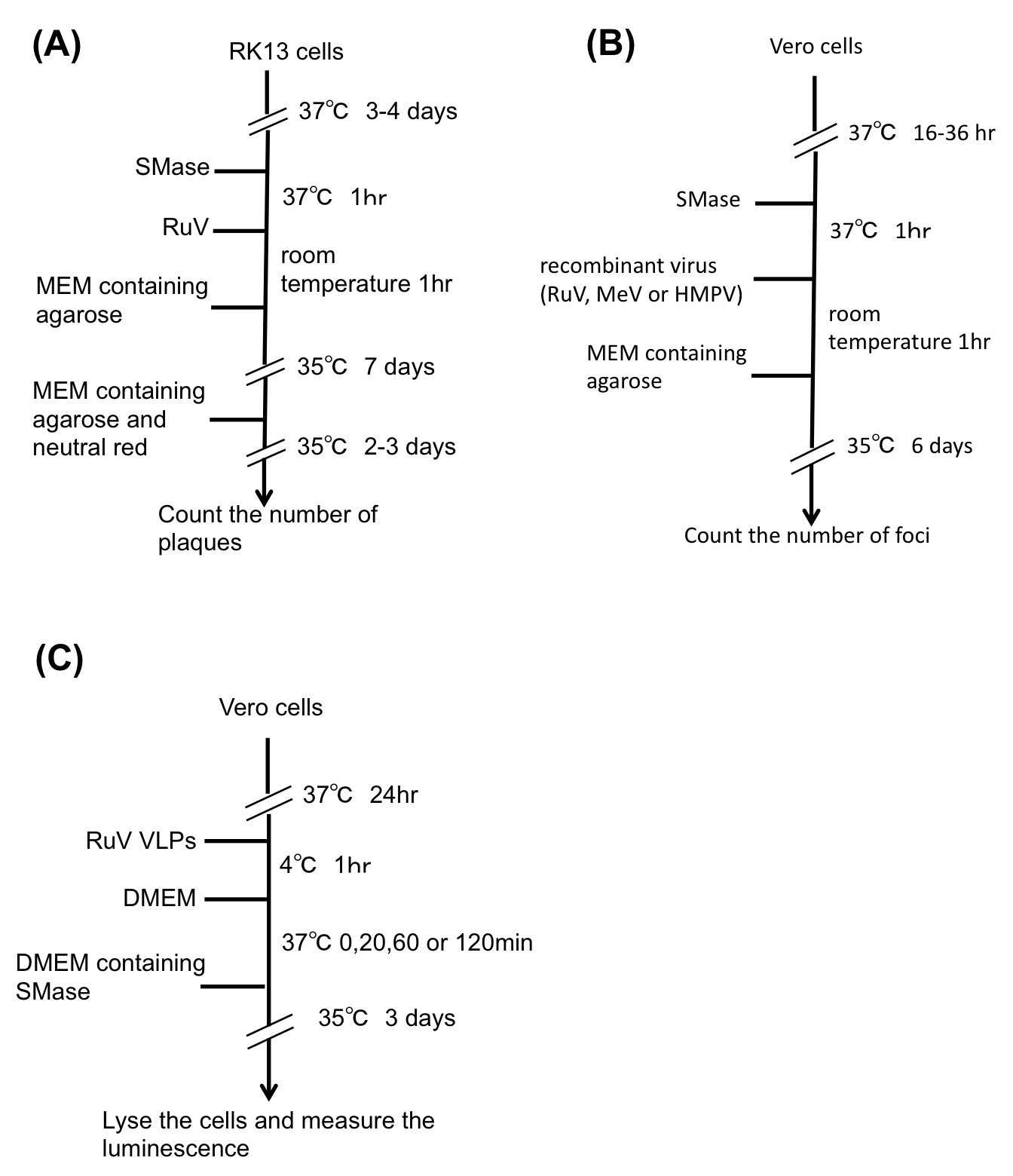
Figure 1. Experimental flow. A. Analysis of the effect of SMase on viral infectivity in RK13 cells. B. Analysis of the effect of SMase on viral infectivity in Vero cells. C. Analysis of the effect of SMase treatment on VLP infection.- Seed 3 ml of a RK13 cell suspension at a concentration of 250,000 cells/ml into each well of a 6-well plate.
- Incubate the cells in a cell culture incubator at 37 °C with 5% CO2.
- After cells form a monolayer (3-4 days post seeding), wash the cells with DPBS once, and then add 1 ml of MEM containing 30 or 150 mU/ml of SMase or 1 ml of MEM as a non-treated control.
Note: The higher concentrations (>150 mU/ml) caused cell damage. The lower concentrations (<30 mU/ml) also showed a dose-dependent effect, but it was difficult to set the perfect experimental condition for reproducing results. - Incubate the cells for 1 h in a cell culture incubator at 37 °C with 5% CO2.
- Wash the cells with DPBS twice. (Remove the DPBS after washing)
- Add 0.1 ml of a RuV-containing solution at a concentration of 300-800 plaque-forming units/ml.
- Incubate the cells with the virus solution for 1 h at room temperature with gentle agitation every 15 min.
- Remove the virus solution, wash the cells with MEM twice to remove unbound viruses. (Remove the MEM after washing)
- Add 3 ml of MEM containing 2% BS and 0.5% agarose.
- Incubate the cells for 7 days in a cell culture incubator at 35 °C with 5% CO2.
- Add 1 ml of MEM containing 0.01% neutral red and 0.5% agarose.
- Incubate for 2 to 3 days in a cell culture incubator at 35 °C with 5% CO2.
- Count the number of plaques with the naked eye (Figure 2).
Note: Usually, plaques are visible in 2 days, but they may become clearer after 3 days incubation.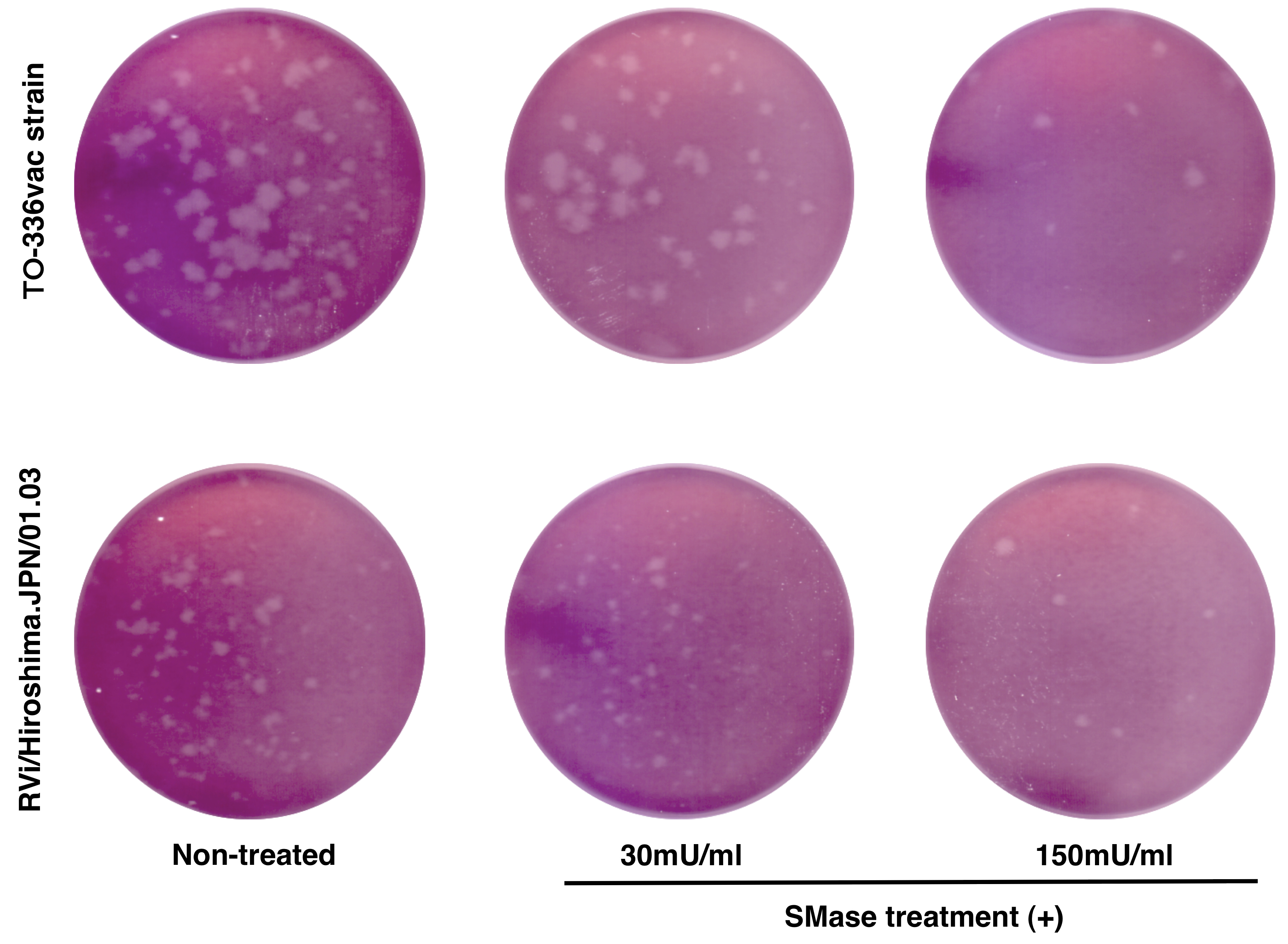
Figure 2. Effect of SMase treatment on RuV infection in RK13 cells. Plaque assays using RK13 cells, which were untreated or treated with 30 or 150 mU/ml of SMase for 1 h before infection. Plaques in cells at 9 days post-infection with the TO-336 vaccine strain (top) or RVi/Hiroshima.JPN/01.03 wild type strain (bottom) visualized by neutral red staining are shown (Otsuki et al., 2018).
- Analysis of the effect of SMase treatment on viral infectivity in Vero cells (Figure 1B).
- Seed 1 ml of a Vero cell suspension at a concentration of 300,000 cells/ml into each well of a 12-well plate.
- Incubate the cells in a cell culture incubator at 37 °C with 5% CO2.
- After cells form a monolayer (16-36 h post seeding), wash the cells with DPBS once, and then add 0.5 ml of DMEM containing 30 or 150 mU/ml of SMase or 0.5 ml of DMEM as a non-treated control. (Remove the DPBS after washing)
- Incubate the cells for 1 h in a cell culture incubator at 37 °C with 5% CO2.
- Wash the cells with DPBS twice. (Remove the DPBS after washing).
- Add 0.1 ml of a virus solution containing RuV rHS/p150-AG1, MeV-IC323/Ed-H-EGFP, or HMPV-rJPS02-76EGFP at a concentration of 300-1000 fluorescent focus-forming units/ml.
- Incubate the cells with the virus solution for 1 h at room temperature with gentle agitation every 20 min.
- Remove the virus solution, wash the cells with MEM twice to remove unbound viruses. (Remove the MEM after washing)
- Add 1 ml of MEM containing 2% FBS and 0.4% agarose.
- Incubate the cells for 6 days in a cell culture incubator at 35 °C with 5% CO2.
- Count the number of foci expressing AG1 or EGFP using a fluorescence microscope (Figure 3).
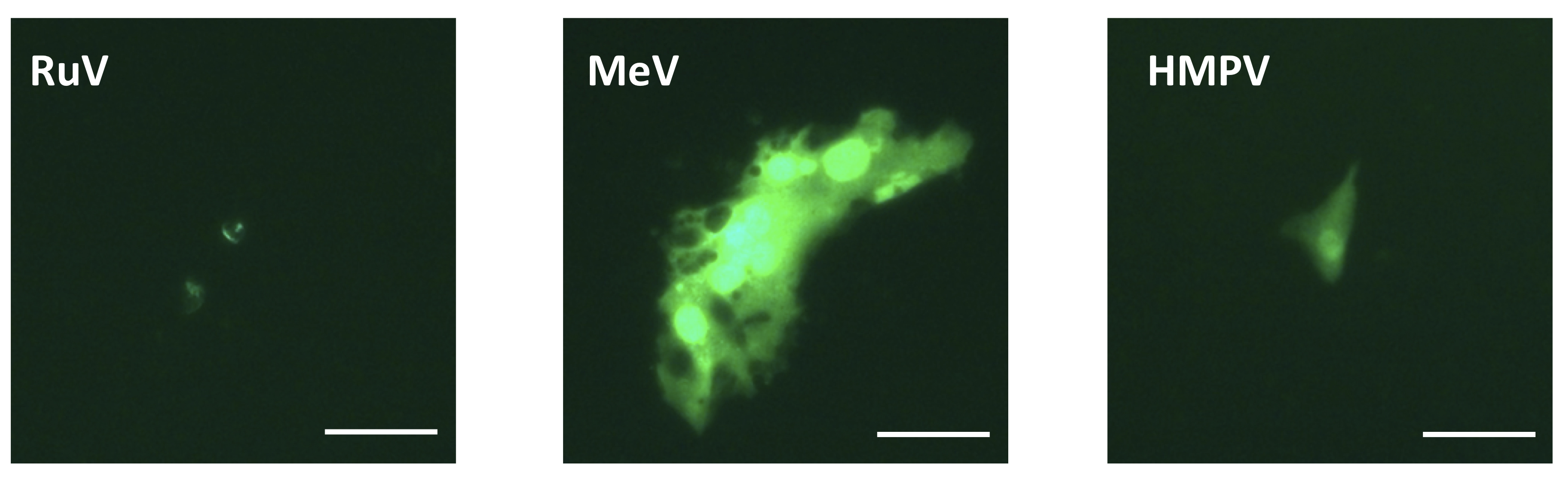
Figure 3. Fluorescence images of Vero cells infected with recombinant viruses. Vero cells were infected with green fluorescent protein-expressing recombinant rubella virus (RuV-rHS/p150-AG1) (left), measles virus (MeV-IC323/Ed-H-EGFP) (center), or human metapneumovirus (HMPV-rJPS02-76EGFP) (right). Images were taken at 6 days post infection. Scale bars represent 50 μm.
- Analysis of the effect of SMase treatment on VLP infection (Figure 1C).
- Seed 0.5 ml of a Vero cell suspension at a concentration of 300,000 cells/ml into each well of a 24-well plate.
- Incubate the cells in a cell culture incubator at 37 °C with 5% CO2.
- After the cells form a monolayer (24 h post seeding), wash the cells with DMEM once. (Remove the DMEM after washing)
- Add 0.1 ml of a RuV-VLP-containing solution at a concentration of 50,000 fluorescent focus-forming units/ml.
- Incubate the cells with the RuV-VLP solution at 4 °C (on ice) for 2 h with gentle agitation every 30 min.
- Wash the cells with DMEM, which is precooled to 4 °C, twice. (Remove the DMEM after washing)
- Add 0.5 ml of DMEM.
- Incubate the cells in a cell culture incubator at 37 °C with 5% CO2.
- Immediately thereafter or after 20, 60 or 120 min, replace the culture media with 0.5 ml of DMEM supplemented with 150 mU/ml of SMase.
- Incubate the cells in a cell culture incubator at 35 °C with 5% CO2 for 3 days.
- Wash the cells with DPBS once and add 50 μl of lysis buffer. (Remove the DPBS after washing)
- Incubate at room temperature for 10 min.
- Transfer the cell lysate to a 1.5 ml tube.
- Prepare 50 μl of Renilla Luciferase Assay Reagent in a new 1.5 ml tube and add 10 μl of the cell lysate into the Renilla Luciferase Assay Reagent-containing tube.
- Mix quickly by flicking the tube with a finger.
- Place the tube in a luminometer to measure the luminescence.
Data analysis
Each assay should be conducted in duplicate to calculate the average number of plaques or foci (or average value of luciferase activity). The assays should be repeated at least three times independently to show statistical differences among the samples.
The percent of virus titer (or luciferase activity) for each assay is calculated as follows:
The percent of virus titer (or luciferase activity) (%) = A/B x100
where, A = average number of plaques or foci (or average value of luciferase activity),
B = average number of plaques or foci (or average value of luciferase activity) in the non-treated control.
Notes
Foci of RuV rHS/p150-AG1 in Vero cells should be carefully counted because the foci of RuV rHS/p150-AG1 are small and produce a weak fluorescence (Figure 3).
Recipes
- MEM
- Dissolve 9.4 g of Eagle’s MEM “Nissui” 1 in 1,000 ml of dH2O, sterilize by using an autoclave
- Then cool at room temperature
- Add penicillin, streptomycin, L-Glutamine, and sodium bicarbonate at the final concentrations of 100 unit/ml, 100 μg/ml, 2 mM, and 0.11%, respectively
- 2x MEM
- Dissolve 9.4 g of Eagle’s MEM “Nissui” 1 in 500 ml of dH2O, sterilize by using an autoclave, and then cool at room temperature
- Add penicillin, streptomycin, L-Glutamine, and sodium bicarbonate at the final concentrations of 200 units/ml, 200 μg/ml, 4 mM, and 0.22%, respectively
- MEM containing 2% BS (or FBS) and 0.5% (or 0.4%) agarose
- Melt 1% (or 0.8%) agarose in dH2O using an autoclave and cool in a water bath set at 40 °C
- Warm 2x MEM containing 4% BS (or FBS) to 40 °C in a water bath as well
- Mix 1% (or 0.8%) agarose solution with the same volume of the 2x MEM containing 4% BS (or FBS), and use immediately, or keep in a water bath at 40 °C
- MEM containing 0.01% neutral red and 0.5% agarose
- Melt 1% agarose in dH2O using an autoclave and cool in a water bath set at 40 °C
- Warm 2x MEM containing 4% BS and 0.02% neutral red to 40 °C in a water bath as well
- Mix the 1% agarose solution with the same volume of the 2x MEM containing 4% BS and 0.02% neutral red, and use immediately, or keep in a water bath at 40 °C
Acknowledgments
We thank Drs. K. Hanada and K. Saito, Department of Biochemistry and Cell Biology, for their invaluable comments. We are also grateful to Ms. M. Nagai for her technical support.
The protocol was developed for our previous work published in Otsuki et al. (2018) supported by AMED-CREST under Grant Number JP18gm0910005 and JSPS KAKENHI (JP15K08508).
Competing interests
We declare that we have no conflict of interest.
References
- Cong, H., Jiang, Y. and Tien, P. (2011). Identification of the myelin oligodendrocyte glycoprotein as a cellular receptor for rubella virus. J Virol 85(21): 11038-11047.
- Mastromarino, P., Cioe, L., Rieti, S. and Orsi, N. (1990). Role of membrane phospholipids and glycolipids in the Vero cell surface receptor for rubella virus. Med Microbiol Immunol 179(2): 105-114.
- Mastromarino, P., Rieti, S., Cioe, L. and Orsi, N. (1989). Binding sites for rubella virus on erythrocyte membrane. Arch Virol 107(1-2): 15-26.
- Ono, N., Tatsuo, H., Hidaka, Y., Aoki, T., Minagawa, H. and Yanagi, Y. (2001). Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol 75(9): 4399-4401.
- Otsuki, N., Abo, H., Kubota, T., Mori, Y., Umino, Y., Okamoto, K., Takeda, M. and Komase, K. (2011). Elucidation of the full genetic information of Japanese rubella vaccines and the genetic changes associated with in vitro and in vivo vaccine virus phenotypes. Vaccine 29(10): 1863-1873.
- Otsuki, N., Sakata, M., Saito, K., Okamoto, K., Mori, Y., Hanada, K. and Takeda, M. (2018). Both sphingomyelin and cholesterol in the host cell membrane are essential for rubella virus entry. J Virol 92(1): JVI.01130-01117.
- Sakata, M., Otsuki, N., Okamoto, K., Anraku, M., Nagai, M., Takeda, M. and Mori, Y. (2014). Short self-interacting N-terminal region of rubella virus capsid protein is essential for cooperative actions of capsid and nonstructural p150 proteins. J Virol 88(19): 11187-11198.
- Seki, F., Takeda, M., Minagawa, H. and Yanagi, Y. (2006). Recombinant wild-type measles virus containing a single N481Y substitution in its haemagglutinin cannot use receptor CD46 as efficiently as that having the haemagglutinin of the Edmonston laboratory strain. J Gen Virol 87(Pt 6): 1643-1648.
- Shirogane, Y., Takeda, M., Iwasaki, M., Ishiguro, N., Takeuchi, H., Nakatsu, Y., Tahara, M., Kikuta, H. and Yanagi, Y. (2008). Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 82(17): 8942-8946.
- Trinh, Q. D., Pham, N. T. K., Takada, K., Komine-Aizawa, S. and Hayakawa, S. (2018). Myelin oligodendrocyte glycoprotein-independent rubella infection of keratinocytes and resistance of first-trimester trophoblast cells to rubella virus in vitro. Viruses 10(1): v10010023.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Otsuki, N., Sakata, M., Mori, Y., Okamoto, K. and Takeda, M. (2018). Analysis of the Effect of Sphingomyelinase on Rubella Virus Infectivity in Two Cell Lines. Bio-protocol 8(17): e2992. DOI: 10.21769/BioProtoc.2992.
Category
Microbiology > Antimicrobial assay > Antiviral assay
Cell Biology > Cell-based analysis > Viral infection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link