- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Liposome Flotation Assay for Studying Interactions Between Rubella Virus Particles and Lipid Membranes
Published: Vol 8, Iss 16, Aug 20, 2018 DOI: 10.21769/BioProtoc.2983 Views: 10011
Reviewed by: Vamseedhar RayaproluBalasubramanian VenkatakrishnanShweta Kailasan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1877 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2457 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3033 Views
Abstract
Rubella virus (RuV) is an enveloped, positive-sense single-stranded RNA virus that is pathogenic to humans. RuV binds to the target cell via the viral envelope protein E1, but the specific receptor molecules on the target cell are yet to be fully elucidated. Here, we describe a protocol for liposome flotation assay to study direct interactions between RuV particles and lipid membranes in a qualitative manner. Interactions are examined by a Nycodenz density gradient fractionation using UV-inactivated RuV particles and fluorescent-labeled liposomes consisting of pure lipids. Fractionated RuV particles are detected using standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis for viral proteins. On the Nycodenz gradient, RuV particles bound to liposomes shift to lower density fractions than unbound RuV particles. Using this protocol, we provide compelling evidence that, at neutral pH in a calcium-dependent manner, RuV particles bind to lipid membranes containing both sphingomyelin (SM) and cholesterol in certain cell types.
Keywords: Liposome flotation assayBackground
Rubella virus is the causative agent of ‘rubella’, an acute and relatively mild systemic infection and ‘congenital rubella syndrome’, a trans-placental fetal infection leading to serious birth defects (Hobman, 2013). Elucidation of molecular mechanisms of RuV entry is essential for understanding viral pathology and helpful for developing anti-RuV drugs. Though previous studies have suggested that membrane lipids of host cells serve as RuV receptors (Mastromarino et al., 1989 and 1990; DuBois et al., 2013), the detailed mechanism remains unknown. Recently, we found that RuV binds to erythrocytes and lymphoid cells in a calcium-dependent manner, and that the calcium-dependent viral binding is impaired after treatment of these cells with sphingomyelinase or cholesterol-adsorbent methyl-β-cyclodextrin, suggesting that SM and cholesterol of the host plasma membrane are critical for binding (Otsuki et al., 2018). To obtain compelling biochemical evidence, we established an assay system to detect interactions between RuV particles and lipids.
Representative biochemical assays widely applied for studying interactions between proteins and lipids are liposome co-sedimentation and co-flotation assays (Zhao and Lappalainen, 2012). Provided that RuV particles and liposomes form aggregates pelleted by low-speed centrifugation in analogy with viral hemagglutination, we initially tried to apply liposome co-sedimentation assay. Unfortunately, our trial of the co-sedimentation assay showed that only a small amount of RuV particles was pelleted at 15,000 x g in the presence of any liposomes. Nevertheless, RuV particles tended to be less pelleted in the presence of liposomes containing both SM and cholesterol, compared with those containing either or neither of the two lipids, providing us with direction for the study. Following this, we devised a flotation assay that can be performed on a small scale. For this, we employed a protocol originally applied for characterization of phosphoinositide binding of the S. cerevisiae Hsv2 (homologous with swollen vacuole phenotype 2) protein (Busse et al., 2013). After making several modifications in the original protocol to optimize for RuV analysis, we have established the protocol described below. By analysis with this protocol, we revealed that both SM and cholesterol are responsible for the calcium-dependent membrane binding of RuV particles.
Materials and Reagents
- Round-bottom glass tubes (size: 16 x 100 mm) (AGC Techno Glass, IWAKI, catalog number: TST-SCR16-100 ) with screw caps (AGC Techno Glass, IWAKI, catalog number: 9998CAP415-15 )
- Round-bottom glass tubes (size: 12 x 75 mm) (AGC Techno Glass, IWAKI, catalog number: 9831-1207 )
- Polypropylene centrifuge tubes:
15 ml (AS ONE, VIOLAMO, catalog number: 1-3500-21 )
50 ml (Corning, Centristar, catalog number: 430829 ) - Polypropylene microfuge tubes:
1.5 ml (FUKAE KASEI, Watson, catalog number: 131-415C )
1.5 ml (Safe-Lock tubes, Eppendorf, catalog number: 0030 120.086 , for heating SDS-PAGE samples)
2.0 ml (FUKAE KASEI, Watson, catalog number: 132-620C ) - Polycarbonate ultracentrifuge tubes (Beckman Coulter, catalog number: 343778 )
- Pipette tips (Quality Scientific Plastics):
1-200 μl (Thermo Fisher Scientific, catalog number: 110-96RSNEW )
100-1,000 μl (Thermo Fisher Scientific, catalog number: 111-NXL-R100S ) - Serological pipets (Costar stripette):
5 ml (Corning, catalog number: 4487 )
10 ml (Corning, catalog number: 4488 ) - Immune-Blot polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories, catalog number: 1620177 )
- Chromatography papers (Whatman 3MM Chr, GE Healthcare, catalog number: 3030-672 )
- Polystyrene containers (180 x 90 x 45 mm) (AS ONE, catalog number: 1-4698-09 )
- Black 96-well strip plate (Black Combiplate 8, Labsystems, catalog number: 95029450 )
Note: This product has been discontinued. - Parafilm (Bemis, catalog number: PM996 )
- 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) (50 mg/ml in chloroform, Avanti Polar Lipids, catalog number: 850375C )
Note: Store at -30 °C. - SM from egg (Avanti Polar Lipids, catalog number: 860061P )
Note: Store at -30 °C. - Cholesterol (Sigma-Aldrich, catalog number: C8667 )
Note: Store at -30 °C. - L-α-Phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl) (Ammonium Salt) (Rhod PE) (1 mg/ml in chloroform, Avanti Polar Lipids, catalog number: 810146C )
Note: Transfer the solution to a round-bottom glass tube with a screw cap. Seal the cap with parafilm to avoid evaporation. Protect from light and store at -30 °C. Approximate molar concentration calculated with molecular weight of predominant species (1275.678 g/mol): 0.8 mM. - Chloroform (Wako Pure Chemical Industries, catalog number: 038-02601 )
Caution: Chloroform is volatile and hepatotoxic. Thus, when using chloroform and its mixtures, one MUST deal with them in a fume hood or with alternative equipment for chemical safety. - Methanol (Sigma-Aldrich, catalog number: 19-2410-4 )
- Ethanol (Wako Pure Chemical Industries, catalog number: 057-00456 )
- Water (ultrapure water, e.g., Milli Q)
- UV-inactivated RuV particles for hemagglutination inhibition test (RuV antigens) (Denka Seiken, catalog number: 310071 )
Note: Store at -30 °C. After reconstitution with 1 ml/vial of water, store at 4 °C and use within 3 days. - Calcium chloride dihydrate (CaCl2•2H2O) (NACALAI TESQUE, catalog number: 06731-05 )
- Tris-buffered saline (10x, pH 7.4) (TBS) (NACALAI TESQUE, catalog number: 35438-81 )
- Protease inhibitor cocktail for use with mammalian cell and tissue extracts (100x) (NACALAI TESQUE, catalog number: 25955-11 )
- 5-(N-2,3-dihydroxypropylacetamido)-2,4,6-triiodo-N,N’-bis(2,3-dihydroxypropyl) isophthalamide (Nycodenz) (Sigma-Aldrich, catalog number: D2158 )
- Sodium dodecyl sulfate (SDS) (NACALAI TESQUE, catalog number: 02873-75 )
- Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
- Dithiothreitol (DTT) (NACALAI TESQUE, catalog number: 14112-94 )
- Bromophenol blue (Wako Pure Chemical Industries, catalog number: 029-02912 )
- Tris(hydroxymethyl)aminomethane (Tris) (NACALAI TESQUE, catalog number: 35434-21 )
- Glycine (NACALAI TESQUE, catalog number: 17109-64 )
- Hydrochloric acid (HCl) (Wako Pure Chemical Industries, catalog number: 080-01066 )
- 30% (w/v)-Acrylamide/bis mixed solution (37.5:1) (NACALAI TESQUE, catalog number: 06144-05 )
- Ammonium peroxodisulfate (APS) (NACALAI TESQUE, catalog number: 02627-34 )
- N,N,N',N'-Tetramethylethylenediamine (TEMED) (NACALAI TESQUE, catalog number: 33401-72 )
- Prestained protein markers such as PINK prestained protein ladders (NIPPON Genetics, catalog number: MWP02 ) and ExcelBandTM 3-color regular range protein maker (SMOBIO Technology, catalog number: PM2500 )
- Skim milk (BD, Difco, catalog number: 232100 )
- Polyoxyethylene(20) sorbitan monolaurate (Tween 20) (Wako Pure Chemical Industries, catalog number: 167-11515 )
- Goat anti-RuV virion (strain HPV-77) polyclonal antibody (Acris Antibodies, catalog number: BP1061 )
Note: Store at 4 °C. For long time storage, aliquot and store at -30 °C. - Mouse monoclonal anti-goat/sheep IgG–peroxidase antibody (Sigma-Aldrich, catalog number: A9452 )
Note: Store at 4 °C. For long-time storage, aliquot and store at -30 °C. - Immobilon Western Chemiluminescent HRP Substrate (Merck, catalog number: WBKLS0500 )
- Chloroform/methanol (19/1, v/v)
- 0.2 M CaCl2
- 1 M DTT
- 1% (w/v) bromophenol blue in 50% (v/v) ethanol
- 10% (w/v) SDS
- 1 M Tris-HCl pH 8.8
- 1 M Tris-HCl pH 6.7
- 10% (w/v) APS
- 20% (w/v) Tween 20
Note: Store at 4 °C. Check bacterial or fungal contamination before use. - Lipid stock solutions
- TBS (see Recipe 4)
- 2x TBS (see Recipe 5)
- 80% (w/v) Nycodenz/TBS (see Recipe 6)
- 30% (w/v) Nycodenz/TBS (see Recipe 7)
- 3x SDS sample buffer (see Recipe 8)
Note: Aliquot and store at -30 °C. - 10% (w/v) polyacrylamide separating gel solution (see Recipe 9)
- 5% (w/v) polyacrylamide stacking gel solution (see Recipe 10)
- 10x Tris/Glycine (see Recipe 11)
- Electrode buffer (see Recipe 12)
- Transfer buffer (see Recipe 13)
- TBS-T (see Recipe 14)
- 5% (w/v) skim milk/TBS-T (see Recipe 15)
- 2% (w/v) skim milk/TBS-T (see Recipe 16)
Equipment
- Micropipettes durable against organic solvent dispensing (NICHIRYO, model: Nichipet EX Plus II, catalog numbers: 00-NPLO2-20 , 00-NPLO2-200 , 00-NPLO2-1000 )
- Pipet-Aid XP Pipette Controller (Drummond Scientific, catalog number: 4-040-101-J )
- Vortex mixer (Delta mixer, TAITEC, model: Se-08 )
- Nitrogen evaporator with water bath (Nakajima seisakusho, Co., Ltd., custom-made)
- Fume hood equipped with activated charcoal filters (Dalton, model: DC-183-100 )
- Probe-type ultrasonic processor (Hielscher Ultrasonics, model: UP50H )
- Micro refrigerated centrifuge (e.g., KUBOTA, model: 3520 )
- Electronic balance (Shimadzu, LIBROR, model: EB-340HW ; Mettler-Toledo International, model: ML802/52 )
- Ultracentrifuge (Beckman Coulter, model: OptimaTM TLX with TLS-55 rotor)
- Block heater for microfuge tubes (Nippon Genetics, Fast GeneTM, model: FG-02N )
- pH meter (TOA Electronics, model: HM-30S )
- Microwave oven (TOSHIBA, model: ER-225 )
- Protein electrophoresis apparatus for SDS-PAGE (e.g., BIO CRAFT, model: BE-S28 for wide mini gels)
- Western blot apparatus (e.g., Bio-Rad Laboratories, CriterionTM blotter for wide mini gels, catalog number: 1704070 )
- Power supply (ATTO, model: AE-8450 , for SDS-PAGE; Bio-Rad Laboratories, model: 250/2.5 , for Western blotting)
- Tube rotator (SCINICS, model: RVM-101 )
- Reciprocal (TAITEC, model: Personal-11 ) and/or seesaw (TAITEC, model: Wave-SI ) shakers
- Chemiluminescent imaging system (ATTO, model: WSE-6200H LuminoGraph II)
- Microplate reader (BMG LABTECH, model: FLUOstar Optima )
Software
- ImageSaver 6 for Windows (ATTO Co.) as image acquisition software for WSE-6200H LuminoGraph II
- CS Analyzer 4 for Windows (ATTO Co.) as image analysis software
- Microsoft Excel 2016 (Microsoft Corp.) as spreadsheet software
- GraphPad Prism 7 (Graph Pad Software) as graph drawing software
Procedure
- Preparation of liposomes
Conduct all manipulations at room temperature unless otherwise stated. When using samples containing fluorescent-labeled lipids, protect them from direct light whenever possible throughout the procedure.- For preparation of 2x concentrated liposomes consisting of DOPC/SM/cholesterol/Rhod PE in a molar ratio of 8:2:3:0.05, prepare a stock solution of each lipid in organic solvent and then mix an appropriate volume of each stock solution in a round-bottom glass tube to obtain the final amounts: 1.6 μmol DOPC, 0.4 μmol SM, 0.6 μmol cholesterol, and 10 nmol Rhod PE (Figure 1A). For liposomes of other compositions, alter the amounts of lipid put into the glass tube appropriately (Table 1).
Table 1. Lipid compositions of 2x concentrated liposomes. Each liposome contains an equal amount of phospholipids. DOPC is used as the matrix phospholipid because phosphatidylcholine is the most abundant type of phospholipid in the plasma membrane. The molar ratio of DOPC/SM/cholesterol of liposome A (8/2/3) is the same as that of the liposomes used in the previous study on sphingomyelin-specific toxin lysenin (Ishitsuka and Kobayashi, 2007). Liposome A and A’ are identical in composition.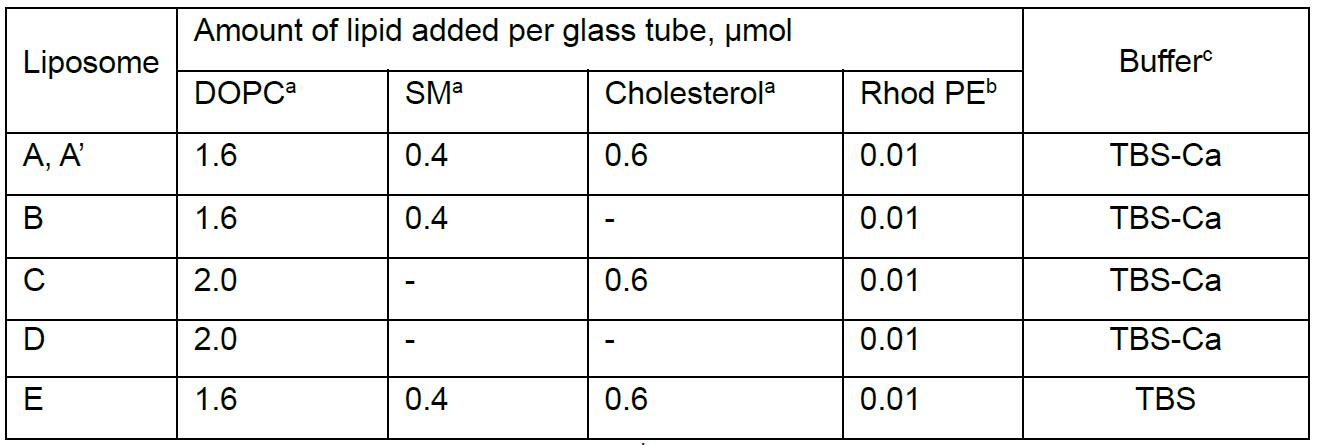
adissolved in chloroform/methanol (19/1, v/v); bdissolved in chloroform; cadded at 1 ml per glass tube after evaporation. - Evaporate organic solvent completely under nitrogen gas at a bath temperature of 35-40 °C using nitrogen evaporator set up in a fume hood. After at least 20 min of evaporation, lipid film is formed at the bottom of the glass tube (Figure 1B).
- Rehydrate the resultant lipid film with 1 ml of TBS-Ca (Figure 1C). To prepare liposomes without calcium ions, use TBS instead of TBS-Ca.
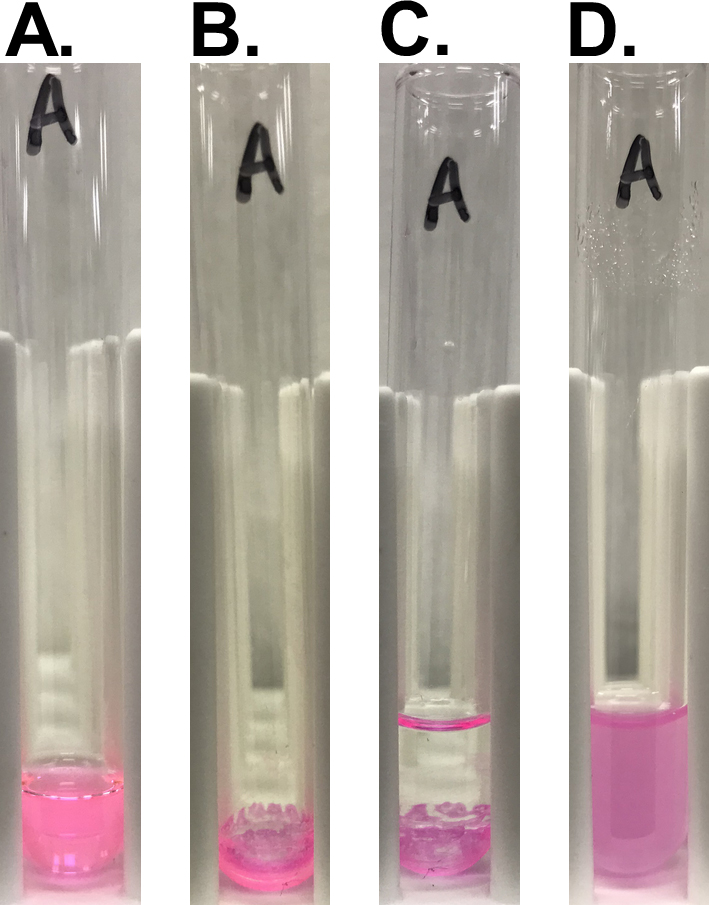
Figure 1. Preparation of liposomes. A. Mixture of lipid solution before evaporation; B. Lipid film formed after evaporation; C. Lipid film rehydrated with TBS-Ca; D. Lipid suspension after sonication. - Sonicate the mixture for 20-30 min with a probe-type sonicator with cooling in tap water (without ice). For UP50H ultrasonic processor, set an amplitude at 80% and a duty cycle at 50%.
Note: The mixture looks turbid at the onset. After 30 min (for liposome A) or 20 min (for liposomes with other compositions) of sonication, the mixture becomes transparent. Vortex the tube every 10 min of sonication to restore liquid spattered on the inner wall of the tube. - Transfer the lipid suspension to a 1.5-ml microfuge tube.
- Centrifuge the lipid suspension at 15,000 x g for 10 min at 25 °C with a micro refrigerated centrifuge to precipitate the lipid aggregates and debris from sonicator tip.
- Transfer the supernatant to a new 1.5 ml microfuge tube and centrifuge again under the same conditions. Repeat centrifugation until no visible pellet is formed.
- Transfer the supernatant to a new 1.5 ml microfuge tube and store the fraction as 2x concentrated liposome fraction at 4 °C. Protect from light and use within 3 days.
- For preparation of 2x concentrated liposomes consisting of DOPC/SM/cholesterol/Rhod PE in a molar ratio of 8:2:3:0.05, prepare a stock solution of each lipid in organic solvent and then mix an appropriate volume of each stock solution in a round-bottom glass tube to obtain the final amounts: 1.6 μmol DOPC, 0.4 μmol SM, 0.6 μmol cholesterol, and 10 nmol Rhod PE (Figure 1A). For liposomes of other compositions, alter the amounts of lipid put into the glass tube appropriately (Table 1).
- Nycodenz density gradient centrifugation
Interaction between RuV antigens and liposomes is assessed using Nycodenz density gradient fractionation. Schematic outline of this experiment is shown in Figure 2. Conduct all manipulations at room temperature unless otherwise stated. Warm all the reagents to room temperature before use. To omit calcium ions from each reaction, use TBS-based reagents instead of TBS-Ca-based ones.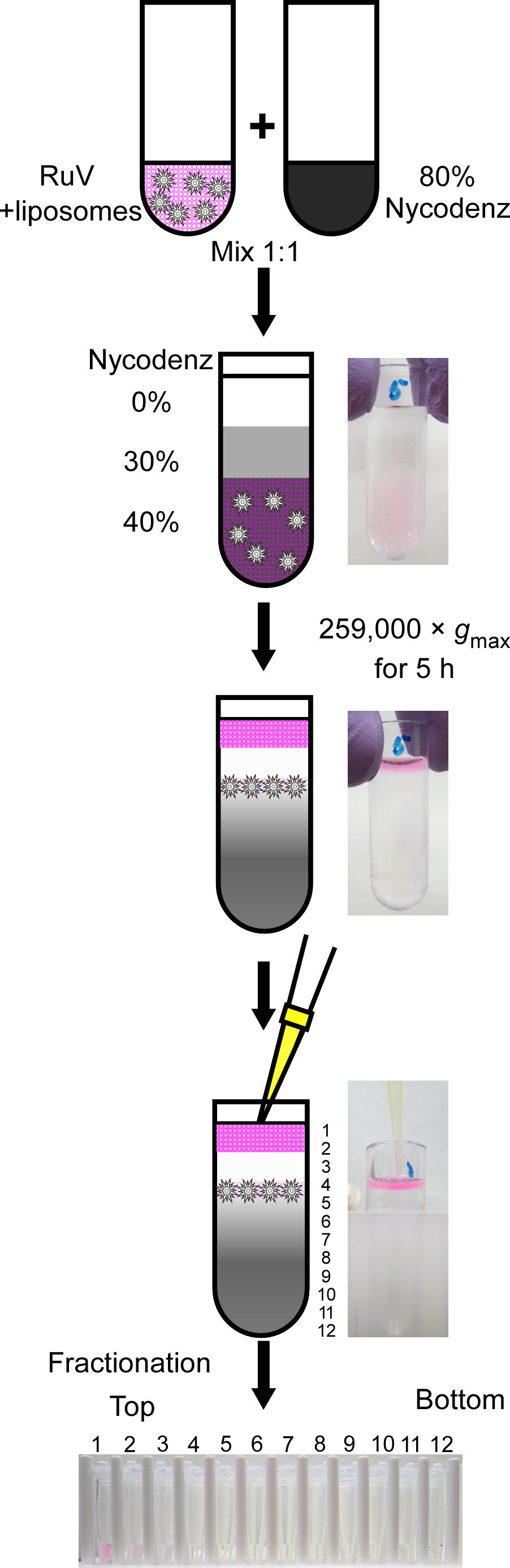
Figure 2. Schematic outline of Nycodenz density gradient fractionation- Mix the following components in a polycarbonate ultracentrifuge tube:
42 μl of Water
75 μl of 2x TBS-Ca
3 μl of 100x protease inhibitor cocktail
30 μl of RuV antigens
(Total volume: 150 μl)
Notes:- RuV antigens must be added at last. Mix well by pipetting before and after the addition of RuV antigens.
- RuV antigens contain bovine serum albumin added as an additive by the manufacturer. Its concentration is not disclosed.
- Add 150 μl of 2x concentrated liposome fraction prepared in Step A8 to the mixture, and mix well by pipetting.
- Incubate the mixture for 1 h, protecting from light.
- Mix the reaction mixture (300 μl) with 300 μl of 80% (w/v) Nycodenz/TBS-Ca by pipetting to obtain homogeneous solution. The resultant solution contains 40% (w/v) Nycodenz.
- Overlay the 40% (w/v) Nycodenz solution with 300 μl of 30% (w/v) Nycodenz/TBS-Ca, being careful not to mix them up.
- Overlay the 30% (w/v) Nycodenz solution with 300 μl of TBS-Ca, being careful not to mix them up.
- Weigh each tube and swinging bucket. Balance weight of two sets of tubes and buckets that will be placed diagonally in TLS-55 rotor in 0.01 g scale by adding TBS-Ca to a tube.
Note: Although the weights of the buckets are initially matched by the manufacturer, the buckets need to be balanced in weight because they can become worn over time. - Set each tube carefully into a swinging bucket.
- Hang the bucket into a TLS-55 rotor carefully, then set the rotor in an Optima TLX ultracentrifuge.
- Centrifuge the samples at 55,000 rpm (259,000 x gmax) for 5 h at 25 °C.
- Remove each tube carefully from the buckets.
- Collect 50 μl aliquots from the top of the gradient using P-200 micropipette with a 1-200 μl tip. Always set the point of the pipette tip just below the aqueous surface (≤ 1 mm) and draw the solution slowly in a circular motion, being careful not to suction air.
- Combine two 50 μl aliquots in a 1.5 ml microfuge tube (i.e., ~100 μl/tube). Although 100 μl aliquots can be taken at once with a micropipette, the authors recommend 50 μl aliquots: A short pipetting stroke is easier to control, thereby lowering the risk of disturbing the gradient. Fractionate each sample into 12 fractions. When the volume of the last fraction is smaller than 100 μl, adjust the volume to ~100 μl with TBS-Ca.
Note: Mark ~100 μl line on 1.5-ml tubes for the last fractions by using 100 μl of water in advance. - Transfer 30 μl of each fraction into a 1.5-ml microfuge tube, then mix with 15 μl of 3x SDS sample buffer. Store at -30 °C until SDS-PAGE analysis. 15 μl of the resultant solution contains 10% (10 μl) of each collected fraction.
- Dilute 24 μl of RuV antigens with 56 μl of TBS-Ca, then mix with 40 μl of 3x SDS sample buffer. Store at -30 °C until SDS-PAGE analysis. 15 μl of the resultant solution contains 3 μl of RuV antigens, which corresponds to 10% of those input into each ultracentrifuge tube. Store the residual fractions (~70 μl/tube) at 4 °C until measurement of fluorescence.
- Mix the following components in a polycarbonate ultracentrifuge tube:
- SDS-PAGE and Western blot analysis
Distribution of RuV particles among the collected fractions is analyzed using standard SDS-PAGE followed by Western blot analysis for RuV proteins. Conduct all manipulations at room temperature unless otherwise stated.- See a basic protocol for SDS-PAGE (He, 2011). Prepare a 10% (w/v) polyacrylamide separating gel using a gel casting device of a protein electrophoresis apparatus according to the manufacturer’s instructions.
- Prepare a 5% (w/v) polyacrylamide stacking gel, similarly.
- Assemble the gel into an electrophoresis device and pour electrode buffer.
- Heat samples for SDS-PAGE analysis prepared in Steps B14 and B15 at 95 °C for 5 min using a block heater, then cool to room temperature. Spin at 15,000 x g for ~10 sec at 25 °C.
- Load 15 μl of the resultant samples per lane of the gel. Load 5 μl of prestained molecular markers similarly.
- Perform electrophoresis at 100-200 V constant according to the manufacturer’s instruction.
- After electrophoresis, place the gel into a clean container and shake it in transfer buffer for 10 min using reciprocal or seesaw shakers. Pre-wet a PVDF membrane with methanol and shake it similarly.
- Set up a sandwich of layers, including chromatography papers, the gel, and the membrane, soaking in transfer buffer.
- Place the sandwich in a chamber of a Western blot apparatus filled with transfer buffer.
- Transfer proteins from the gel to the membrane electrophoretically at 100 V constant for 30 min in a cold room at 4 °C according to the manufacturer’s instructions.
- Place the membrane in a clean container and soak in 5% (w/v) skim milk/TBS-T for 1 h with shaking.
- Wash the membranes three times for 5 min each in TBS-T.
- Place the membrane in a clean container and add the primary antibody (goat anti-RuV polyclonal antibody) diluted 1:4,000 with 2% (w/v) skim milk/TBS-T.
- Incubate the membrane at 4 °C overnight with shaking.
- Wash the membranes three times for 5 min each in TBS-T.
- Place the membrane in a clean container and add the secondary antibody (HRP-conjugated mouse anti-goat/sheep IgG) diluted1:4,000 with 2% (w/v) skim milk/TBS-T.
- Incubate the membrane for 1 h with shaking.
- Wash the membranes three times for 10 min each in TBS-T.
- Develop signal by incubating the membrane with Immobilon Western Chemiluminescent HRP Substrate or an equivalent reagent according to the manufacturer’s instructions.
- Detect the signal using a WSE-6200H LuminoGraph II image analyzer according to the manufacturer’s instructions.
- Measurement of fluorescence
Distribution of liposomes among the collected fractions is analyzed based on fluorescence intensity of Rho PE. Warm the collected fractions and 2x concentrated liposome fractions to room temperature before measurement.- Transfer 50 μl of each collected fraction (which corresponds to 50% of each fraction) and 2x concentrated liposome fraction (which corresponds to 33% of total input) into a well of a 96-well black microplate.
- Measure fluorescence intensity of Rhod PE in each well using a FLUOstar Optima microplate reader at 544 nm excitation and 590 nm emission in endpoint mode without shaking function according to the manufacturer’s instruction.
- Save results in an excel file format.
Data analysis
- Visualize RuV proteins in the collected fraction using image acquisition and analysis software. The apparent molecular weights of RuV E1 (one of two envelope glycoproteins) and capsid proteins are 58-65 kDa and 33-38 kDa, respectively. Figure 3 shows the distribution of E1 and capsid proteins on Nycodenz density gradient fractionation in the presence or absence of various liposomes. RuV particles appear to be present in fractions that contained both proteins. The dense fractions (fractions 10-12) contained only the capsid protein, suggesting that RuV particles were absent from these fractions. Although RuV particles were mainly detected in fraction 6 in the absence of liposomes (Figure 3F), they shifted to fraction 4 in the presence of liposomes consisting of DOPC/SM/cholesterol (Figures 3A and 3A'). This shift toward lower densities was not observed when calcium ions were omitted from the reaction (Figure 3E), indicating that RuV particles interact with lipid membranes in a calcium-dependent manner, consistent with a previous study (Dubé et al., 2014). Furthermore, the shift was no longer observed when either SM or cholesterol was omitted from the liposomes (Figures 3B and 3C) and liposomes consisting of DOPC alone were used (Figure 3D), indicating that the calcium-dependent interaction between RuV particles and lipid membranes requires both SM and cholesterol in the membranes. In Figure 3, results of a separate experiment from that described in the original article (Otsuki et al., 2018) are shown. Similar results were obtained from three independent experiments.
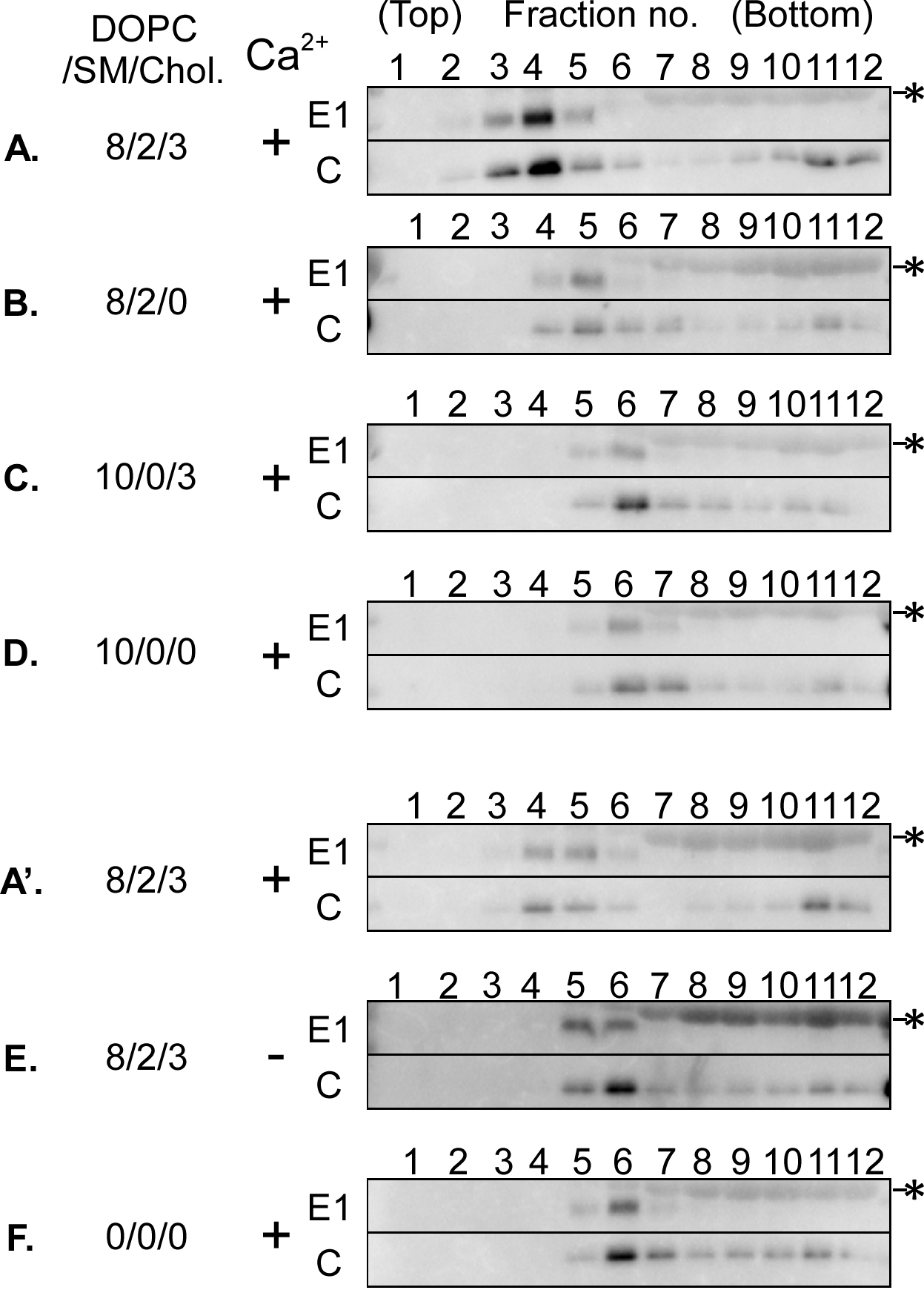
Figure 3. Distribution of RuV proteins on the Nycodenz gradient. RuV antigens (UV-inactivated RuV particles) were incubated with or without various compositions of liposomes in the presence (+) or absence (-) of calcium ions and then subjected to liposome flotation assays on the Nycodenz gradient. Viral E1 and capsid (C) proteins in each fraction were detected by Western blot analysis. An alphabetic code of each panel corresponds to that of liposomes described in Table 1 except for panel F, which indicates the result without liposomes. The values indicated are the amounts (nmol) of DOPC, SM, and cholesterol (Chol.) in each liposome fraction added per μl of RuV antigens. All of the liposomes also contained 0.05 nmol (per μl of RuV antigens) of Rhod PE. Asterisks indicate bovine serum albumin, which was an additive to RuV antigens. Two separate sets of centrifugation experiments are shown: One set, the upper four panels (A-D), and another set, the lower three panels (A'-F). Image acquisition was carried out under the same conditions (10 min of exposure in high sensitivity mode), and contrast of images was adjusted to the same extent (setting upper value at 40,000 and lower value at zero). Panels A and A' represent results from separate experiments using liposomes with the same lipid composition and the same experimental procedures. In this assay, the distribution of viral proteins was similar, but their overall signal intensity varied among the experiments. Though the overall signal intensity was not the main issue of this assay, a possible cause of this variability was an inconstant transfer efficiency in each gel. - Quantify capsid protein using image analysis software and determine recovery of the protein in each fraction. For this purpose, E1 protein is not suitable, because a large amount of bovine serum albumin added as a preservative to RuV antigens overlaps substantially with E1 protein on a Western blot of the input fraction. Total recovery of capsid protein varied from ~10% to ~70% with experiments probably due to the semi-quantitative nature of Western blot analysis and/or degradation of the protein during or after ultracentrifugation. We did not find any correlation between the type of liposome and the recovery of capsid protein.
- To check distribution of liposomes, calculate % of Rhod PE recovered in each fraction using the data of fluorescent intensity. Figure 4 shows the distribution of liposomes among density gradient fractions. The majority of liposomes (~70% or more) were recovered in the top and second fractions irrespective of lipid compositions. Since floating liposomes can not be completely recovered, a small amount of them (< 10%) remained on the aqueous surface throughout the fractionation procedure and was recovered in the last fraction. In Figure 4, results of a separate experiment from that described in the original article (Otsuki et al., 2018) are shown. Similar results were obtained from three independent experiments.
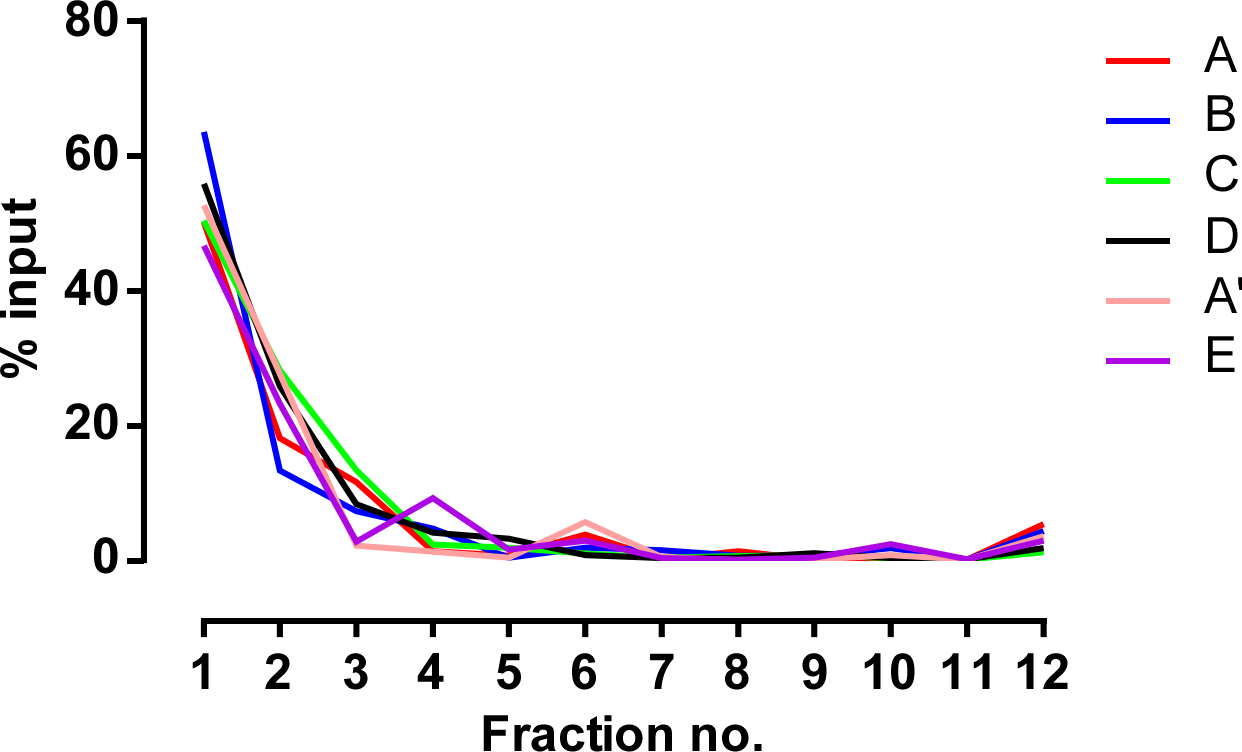
Figure 4. Distribution of liposomes on the Nycodenz gradient. Distribution of liposomes was monitored using the fluorescence intensity of Rhod PE. Values are expressed as a percentage (%) of the total fluorescence intensity of Rhod PE retrieved from each fraction of the gradient. Alphabetic codes correspond to liposomes described in Table 1.
Notes
- Homepage of Avanti Polar Lipids, Inc. (https://avantilipids.com/) is informative for beginners in handling of lipids.
- Homepage of Axis-Shield Density Gradient Media, a brand of Alere Technologies AS (http://www.optiprep.com/) provides general information about Nycodenz.
- Peak positions of RuV particles are mostly reproducible between experiments, but can vary depending on slight differences in the respective experimental conditions. Thus, samples to be directly compared should be prepared, centrifuged, and fractionated in parallel.
Recipes
Note: The water used in the following recipes is ultrapure water unless otherwise stated.
- DOPC stock solution (5 mg/ml)
- Put 1 ml of DOPC (50 mg/ml in chloroform) into a round-bottom glass tube with a screw cap
- Dilute with 9 ml of chloroform/methanol (19/1, v/v)
- Seal the cap with parafilm to lessen evaporation of the solvent
- Store at -30 °C and warm to room temperature before use
- SM stock solution (5 mg/ml)
- Weigh 50 mg of SM (powder) and put into a round-bottom glass tube with a screw cap
- Dissolve SM in 10 ml of chloroform/methanol (19/1, v/v)
- Seal the cap with parafilm to lessen evaporation of the solvent
- Store at -30 °C. Warm to room temperature before use
- Cholesterol stock solution (2 mg/ml, calculated molar concentration: 5.17 mM)
- Weigh 20 mg of cholesterol (powder) and put into a round-bottom glass tube with a screw cap
- Dissolve cholesterol in 10 ml of chloroform/methanol (19/1, v/v)
- Seal the cap with parafilm to lessen evaporation of the solvent
- Store at -30 °C. Warm to room temperature before use
- TBS
25 mM Tris-HCl pH 7.4
137 mM NaCl
2.68 mM KCl- Add 2.5 ml of 10x TBS into a 50-ml centrifuge tube
- Fill up to 25 ml with water and mix well
Store at 4 °C - 2x TBS
Dilute 400 μl of 10x TBS with 1.6 ml of water in a 2-ml microfuge tube
To prepare 2x TBS-Ca, add 20 μl of 0.2 M CaCl2 (final concentration: 2 mM) and mix well
Store at 4 °C - 80% (w/v) Nycodenz/TBS
- Dissolve 8 g of Nycodenz with hot water in a 15-ml centrifuge tube
Note: It requires time to dissolve such an amount of Nycodenz in less than 10 ml of water. Use hot water preheated by a microwave and mix intensively. Warm the tube occasionally with hot tap water. If hard-to-melt clumps appear, disrupt them by sonication in a bath-type sonicator. - Add 1 ml of 10x TBS and fill up to 10 ml with water. Mix well and store at 4 °C.
- Before use, transfer an appropriate amount of the solution into a 2.0-ml microfuge tube and add 1/100 volume of 100x protease inhibitor cocktail. Mix well.
- Dissolve 8 g of Nycodenz with hot water in a 15-ml centrifuge tube
- 30% (w/v) Nycodenz/TBS
- Dissolve 3 g of Nycodenz in hot water in a 15-ml centrifuge tube
- Add 1 ml of 10x TBS
- Fill up to 10 ml with water and mix well. Store at 4 °C
- Before use, transfer an appropriate amount of the solution into a 2.0-ml microfuge tube and add 1/100 volume of 100x protease inhibitor cocktail. Mix well
- 3x SDS sample buffer
- Mix the following components in a 15-ml centrifuge tube:
4.5 ml of Glycerol (final concentration: 30% [v/v])
2.25 ml of 1 M DTT (final concentration: 150 mM)
1.5 ml of 1% (w/v) bromophenol blue in 50% (v/v) ethanol (final concentration: 0.1% [w/v])
2.81 ml of 1 M Tris-HCl pH 6.7 (final concentration: 187.5 mM) - Dissolve 0.9 g of SDS (final concentration: 6% [w/v]) using a tube rotator
- Fill up to 15 ml with water and mix well
- Store at -30 °C and warm at 37 °C to dissolve SDS before use
- Mix the following components in a 15-ml centrifuge tube:
- 10% (w/v) polyacrylamide separating gel solution
- Mix the following components in a 50-ml centrifuge tube:
8.28 ml of Water
11.25 ml of 1 M Tris-HCl pH 8.8 (final concentration: 375 mM)
10 ml of 30% (w/v)-Acrylamide/bis mixed solution
300 μl of 10% (w/v) SDS (final concentration: 0.1% [w/v])
300 μl of 10% (w/v) APS (final concentration: 0.1% [w/v]) - Add 15 μ of TEMED (final concentration: 0.05% [v/v]) and mix well
- Pour into gel casting apparatus immediately
- Mix the following components in a 50-ml centrifuge tube:
- 5% (w/v) polyacrylamide stacking gel solution
- Mix the following components in a 15-ml centrifuge tube:
6.92 ml of Water
1.25 ml of 1 M Tris-HCl pH 6.7 (final concentration: 125 mM)
1.67 ml of 30% (w/v)-Acrylamide/bis mixed solution
100 μl of 10% (w/v) SDS (final concentration: 0.1% [w/v])
100 μl of 10% (w/v) APS (final concentration: 0.1% [w/v]) - Add 10 μ of TEMED (final concentration: 0.1% [v/v]) and mix well
- Cast immediately on separating gels
- Mix the following components in a 15-ml centrifuge tube:
- 10x Tris/Glycine
- Dissolve 121 g of Tris and 576 g of glycine in about 3 L of water
- Fill up to 4 L with water and mix well
- Store at room temperature
- Electrode buffer
- Dilute 100 ml of 10x Tris/Glycine with about 800 ml of water
- Add 10 ml of 10% (w/v) SDS
- Fill up to 1 L with water and mix well
- Store at room temperature
- Transfer buffer
- Dilute 200 ml of 10x Tris/Glycine with 1.4 L of water
- Add 400 ml of methanol and mix well
- Keep on ice or at 4 °C
- TBS-T
- Dilute 100 ml of 10x TBS with less than 900 ml of water
- Add 5 ml of 20% (w/v) Tween 20 to give a final concentration of 0.1% (w/v)
- Fill up to 1 L with water and mix well
- Store at 4 °C and warm to room temperature before use
- 5% (w/v) skim milk/TBS-T
Dissolve 2.5 g of skim milk in 50 ml of TBS-T in a 50 ml centrifuge tube using a tube rotator
Note: Mix the tube for at least 1 h for complete dissolving. Prepare on the day of use and store at 4 °C. Use within 2 days. - 2% (w/v) skim milk/TBS-T
Dissolve 1 g of skim milk in 50 ml of TBS-T in a 50 ml centrifuge tube using a tube rotator
Note: Mix the tube for at least 1 h for complete dissolving. Prepare on the day of use and store at 4 °C. Use within 2 days.
Acknowledgments
This protocol has been used in Otsuki et al. (2018). This study was supported by a grant to KH from AMED-CREST (Japan Agency for Medical Research and Development, Core Research for Evolutional Science and Technology, grant number: JP18gm0910005). This protocol was adapted from the work by Busse et al. (2013).
Competing interests
The authors declare that they have no conflict of interest.
References
- Busse, R. A., Scacioc, A., Hernandez, J. M., Krick, R., Stephan, M., Janshoff, A., Thumm, M. and Kuhnel, K. (2013). Qualitative and quantitative characterization of protein-phosphoinositide interactions with liposome-based methods. Autophagy 9(5): 770-777.
- Dubé, M., Rey, F. A. and Kielian, M. (2014). Rubella virus: first calcium-requiring viral fusion protein. PLoS Pathog 10(12): e1004530.
- DuBois, R. M., Vaney, M. C., Tortorici, M. A., Kurdi, R. A., Barba-Spaeth, G., Krey, T. and Rey, F. A. (2013). Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature 493(7433): 552-556.
- He, F. (2011). Laemmli-SDS-PAGE. Bio-protocol Bio101: e80.
- Hobman, T. C. (2013). Rubella virus. In: Knipe, D. M., Howley, P. M., Cohen, J. I., Griffin, D. E., Lamb, R. A., Martin, M. A., Racaniello, V. R., Roizman, B. (Eds.). Fields virology. 6th edition. vol 1. p 687-711. Lippincott Williams & Wilkins, Philadelphia, PA.
- Ishitsuka, R. and Kobayashi, T. (2007). Cholesterol and lipid/protein ratio control the oligomerization of a sphingomyelin-specific toxin, lysenin. Biochemistry 46(6): 1495-1502.
- Mastromarino, P., Cioè, L., Rieti, S. and Orsi, N. (1990). Role of membrane phospholipids and glycolipids in the Vero cell surface receptor for rubella virus. Med Microbiol Immunol 179(2): 105-114.
- Mastromarino, P., Rieti, S., Cioè, L. and Orsi, N. (1989). Binding sites for rubella virus on erythrocyte membrane. Arch Virol 107(1-2): 15-26.
- Otsuki, N., Sakata, M., Saito, K., Okamoto, K., Mori, Y., Hanada, K. and Takeda, M. (2018). Both sphingomyelin and cholesterol in the host cell membrane are essential for Rubella virus entry. J Virol. 92(1): e01130-17.
- Rouser, G., Siakotos, A. N. and Fleischer, S. (1966). Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids 1(1): 85-86.
- Zhao, H. and Lappalainen, P. (2012). A simple guide to biochemical approaches for analyzing protein-lipid interactions. Mol Biol Cell 23(15): 2823-2830.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Saito, K., Otsuki, N., Takeda, M. and Hanada, K. (2018). Liposome Flotation Assay for Studying Interactions Between Rubella Virus Particles and Lipid Membranes. Bio-protocol 8(16): e2983. DOI: 10.21769/BioProtoc.2983.
Category
Microbiology > Microbe-host interactions > Virus
Biochemistry > Lipid > Lipid-virus interaction
Microbiology > Microbe-host interactions > In vitro model
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










