- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Direct Visualization of the Multicopy Chromosomes in Cyanobacterium Synechococcus elongatus PCC 7942
Published: Vol 8, Iss 15, Aug 5, 2018 DOI: 10.21769/BioProtoc.2958 Views: 6923
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Super-resolution Microscopy at Cryogenic Temperatures Using Solid Immersion Lenses
Benji C. Bateman [...] Marisa L. Martin-Fernandez
Nov 20, 2019 6728 Views

Attachment of a 32P-phosphate to the 3′ Terminus of a DNA Oligonucleotide
Joshua C. Cofsky and Jennifer A. Doudna
Oct 20, 2020 5157 Views

Calibrating Fluorescence Microscopy With 3D-Speckler (3D Fluorescence Speckle Analyzer)
Chieh-Chang Lin and Aussie Suzuki
Aug 20, 2024 1950 Views
Abstract
Cyanobacteria are prokaryotic organisms that carry out oxygenic photosynthesis. The fresh water cyanobacterium Synechococcus elongatus PCC 7942 is a model organism for the study of photosynthesis and gene regulation, and for biotechnological applications. Besides several freshwater cyanobacteria, S. elongatus 7942 also contains multiple chromosomal copies per cell at all stages of its cell cycle. Here, we describe a method for the direct visualization of multicopy chromosomes in S. elongatus 7942 by fluorescence in situ hybridization (FISH).
Keywords: CyanobacteriaBackground
Cyanobacteria are prokaryotic microorganisms that utilize an oxygen-producing photosynthetic system similar to that of chloroplasts in higher plants. Whereas bacteria such as Escherichia coli and Bacillus subtilis harbor a single circular chromosome, several species of freshwater cyanobacteria have multiple circular chromosomes per cell (Mann and Carr, 1974; Labarre et al., 1989; Binder and Chisholm, 1995). The freshwater cyanobacterium Synechococcus elongatus PCC 7942 (hereafter referred to as S. elongatus 7942) carries 3-8 chromosomal copies per cell (Griese et al., 2011; Watanabe et al., 2012; Watanabe et al., 2015). A fluorescence reporter-operator system has been established to acquire images of the multicopy chromosome of S. elongatus 7942 (Chen et al., 2012; Jain et al., 2012). Although this system enables live-cell imaging, it requires complex genetic constructs for labeling individual chromosomes, and, overall, this type of system is unstable over an extended period of cultivation. Thus, it is necessary to examine the chromosomal distribution in S. elongatus 7942 cells that have a simple genetic background. We recently established a fluorescence in situ hybridization (FISH) method for visualizing the oriC region and putative terC region of the multicopy chromosome in S. elongatus 7942 (Watanabe et al., 2018). A DNA probe, which covers the oriC or terC region, was synthesized and used for the FISH analysis. Labeled chromosomes in S. elongatus 7942 cells can be observed by fluorescence microscopy. Here, we describe a step-by-step protocol for the FISH method.
Materials and Reagents
- Pipette tips
200 μl (FUKAEKASEI and WATSON, catalog number: 110-705C )
1,000 μl (FUKAEKASEI and WATSON, catalog number: 110-706C ) - PCR-tubes and strips
Tube (FUKAEKASEI and WATSON, catalog number: 137-333C )
Cap (FUKAEKASEI and WATSON, catalog number: 137-432C ) - Disposable plastic dish (for BG-11 plate) (KANTO KAGAKU, model: CSPD90-15, catalog number: 96930-01 )
- 93 ml test tubes (for culturing) (MonotaRO, IWAKI, catalog number: 09184061)
Manufacturer: IWAKI, catalog number: TEST30NP . - Poly-lysine coated glass slides (Matsunami Glass, catalog number: S7441 )
- Cover glass (MonotaRO, catalog number: CT18189 )
- Tube for sonication (microtube AFA Fiber Pre-Slit Snap-Cap) (Covaris, catalog number: 520077 )
- Aluminum foil
- Synechococcus elongatus PCC 7942 strain (Pasteur culture collection)
- Genomic DNA of S. elongatus 7942 (10-50 ng for each PCR reaction)
- ExTaq PCR Kit (TaKaRa Bio, catalog number: RR001A )
- Fluorescence-12-dUTP (Roche Diagnostics, catalog number: 11427857910 )
- Random primed DNA labeling kit (Roche Diagnostics, catalog number: 11004760001 )
- 99.5% ethanol (KANTO KAGAKU, catalog number: 14032-08 )
- 3 M NaOAc solution (pH 5.2) (KANTO KAGAKU, catalog number: 37092-00 )
- Glycogen (20 ng/ml) (Roche Diagnostics, catalog number: 10901393001 )
- Triton X-100 (NACALAI TESQUE, catalog number: 35501-02 )
- Formamide (KANTO KAGAKU, catalog number: 16062-00 )
- Paraformaldehyde (KANTO KAGAKU, catalog number: 32034-12 )
- Dimethyl sulfoxide (KANTO KAGAKU, catalog number: 10378-00 )
- Sodium hydroxide (KANTO KAGAKU, catalog number: 37184-00 )
- Methanol (KANTO KAGAKU, catalog number: 25183-00 )
- PBS powder (Sigma-Aldrich, catalog number: P3813 )
- Lysozyme (Wako Pure Chemical Industries, catalog number: 122-02673 )
- Tris-HCl (pH 7.5) (Sigma-Aldrich, catalog number: T3253-250G )
- EDTA (NACALAI TESQUE, catalog number: 15111-45 )
- Sodium chloride (KANTO KAGAKU, catalog number: 37144-00 )
- Trisodium citrate dihydrate (KANTO KAGAKU, catalog number: 37150-00 )
- Glycerol (KANTO KAGAKU, catalog number: 17029-00 )
- DAPI (Sigma-Aldrich, catalog number: D9564-10MG )
- Salmon sperm DNA (Roche Diagnostics, catalog number: 11467140001 )
- Three sets of PCR primers for visualizing oriC or terC (Eurofins Genomics, Table 1)
Table 1. List of primers used for the preparation of FISH probes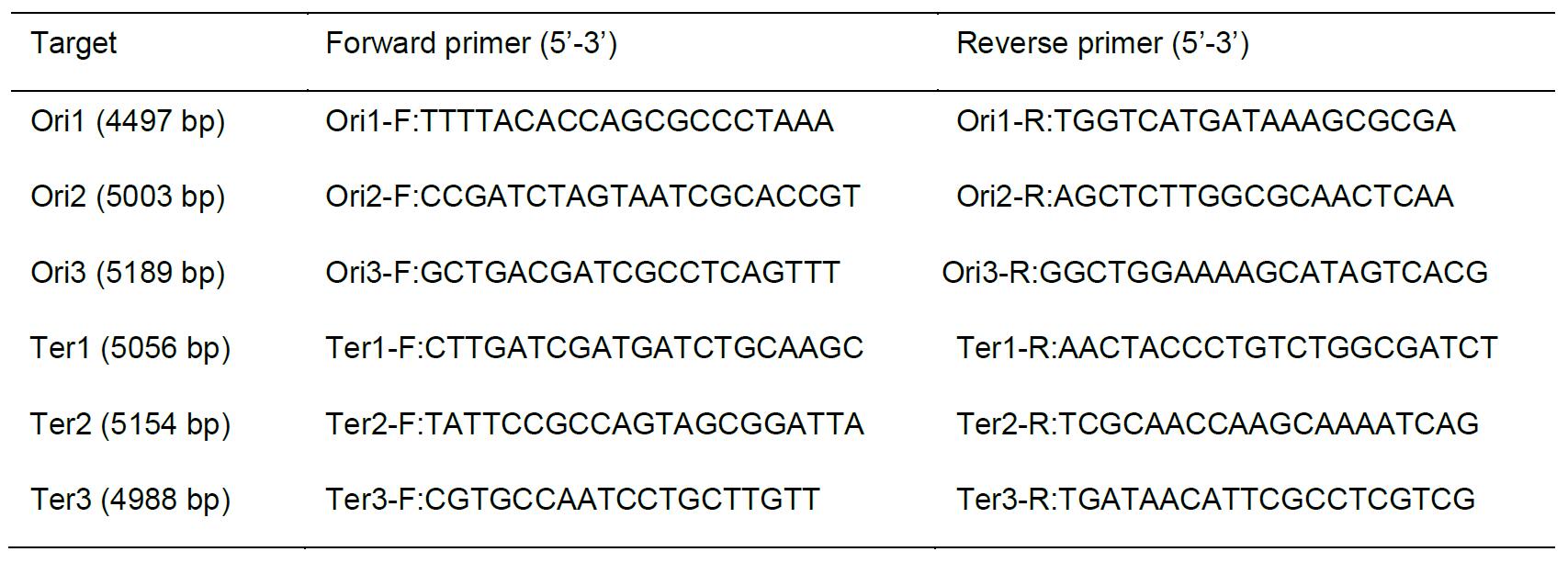
- BG-11 liquid medium (pH 8.2) (Castenholz, 1988)
- BG-11 plates (pH 8.2) (Castenholz, 1988)
- Hybridization solution (see Recipes)
- Fixation solution (see Recipes)
- PBS buffer (see Recipes)
- Lysozyme solution (see Recipes)
- 20x Standard Saline Citrate (SSC) (see Recipes)
- Mounting solution (see Recipes)
Equipment
- Pipettes
P20 (Gilson, catalog number: F123600 )
P200 (Gilson, catalog number: F123601 )
P1000 (Gilson, catalog number: F123602 ) - Thermal cycler (Thermo Fisher Scientific, Applied Biosystems, model: Veriti )
- Microtube centrifuge (TOMY SEIKO, model: MX-107 )
- Covaris S-2 sonicator (Covaris, Woburn, MA, USA)
- Plant growth chamber (TOMY SEIKO, model: CLE-405 )
- UV/Vis spectrophotometer (Shimadzu, model: UV-1800 )
- Pharmaceutical refrigerator (Panasonic, model: MPR-3120CN-PJ )
- Glass vat for staining (AS ONE, models: 1-4398-01 and 1-4398-11 )
- Forma direct heat CO2 Incubator (Thermo Fisher Scientific, model: FormaTM 310 , catalog number: 320)
- Fluorescence microscope (Olympus, models: BX53 and DP71 ; filters: U-FUW for DAPI and U-FGW for chlorophyll, objective: UPLSAPO 100XOPH)
- All-in-one fluorescence microscope (Olympus, model: FSX100 , filters: U-MNUA2 for DAPI, U-MWIBA3 for FISH signal, and U-MWIG3 for chlorophyll, objective: UPLSAPO , super apochromat)
Note: This system is necessary to obtain clear FISH images.
Software
- Camera software for DP71 (Olympus)
- Software for FSX 100 (Olympus, FSX-BSW)
- Adobe Photoshop Elements 11 (Adobe Photoshop, CC)
Procedure
- Preparation of DNA probes
- To create the probe templates, amplify three DNA fragments covering the sequences encompassing the oriC region (primers Ori1-F and Ori1-R, Ori2-F and Ori2-R, and Ori3-F and Ori3-R, Table 1) and three fragments encompassing the putative terC region (primers Ter1-F and Ter1-R, Ter2-F and Ter2-R, and Ter3-F and Ter3-R, Table 1) by PCR with a thermal cycler using ExTaq (see manufacturer'sinstruction). The amplified sequences will then be used to create probe templates (Figure 1).
- To obtain the fragmented DNA (< 100 bp) sonicate the DNA using a Covaris S-2 sonicator (duty cycle: 5%; intensity: 3; cycles/burst: 200; time: 45 sec; number of cycles: 2) (Figure 1). This fragmentation step is necessary for permeating cell membrane.
- Ethanol precipitation: Add 1/10 volume of sodium acetate (pH 5.2), 1/10 volume of glycogen, and 2.5 volume of ethanol to the reaction mix and chill on ice for 10 min. Centrifuge at 20,000 x g for 20 min and wash the pellet using 500 μl of 70% ethanol.
- Dissolve the pellet in 10-50 μl of sterile water and determine the DNA concentration by using a UV/Vis spectrophotometer.
- Label 1 μg of template DNA fragments using a random-primed DNA labeling kit (see the manufacturer'sinstruction) and fluorescence-12-dUTP as the labeling substrate (Figure 1). Denature the template DNA by heating at 100 °C for 10 min and immediately chill on ice for 10 min. Add 10 μl of hexanucleotide mix, 2.5 μl of fluorescence-12-dUTP, dNTP stock mix, and fill up to 95 μl using sterile water. Start the labeling reaction by addition of 5 μl Klenow enzyme and incubate at 37 °C for 15-20 h.
- Remove any remaining non-incorporated substrate by ethanol precipitation (see Step A3). Add 100 μl of the hybridization solution to dissolve the labeled probe DNA (final conc. 10 ng/μl).
- Immediately before hybridization, denature the probe by heating at 100 °C for 10 min and then chill on ice.
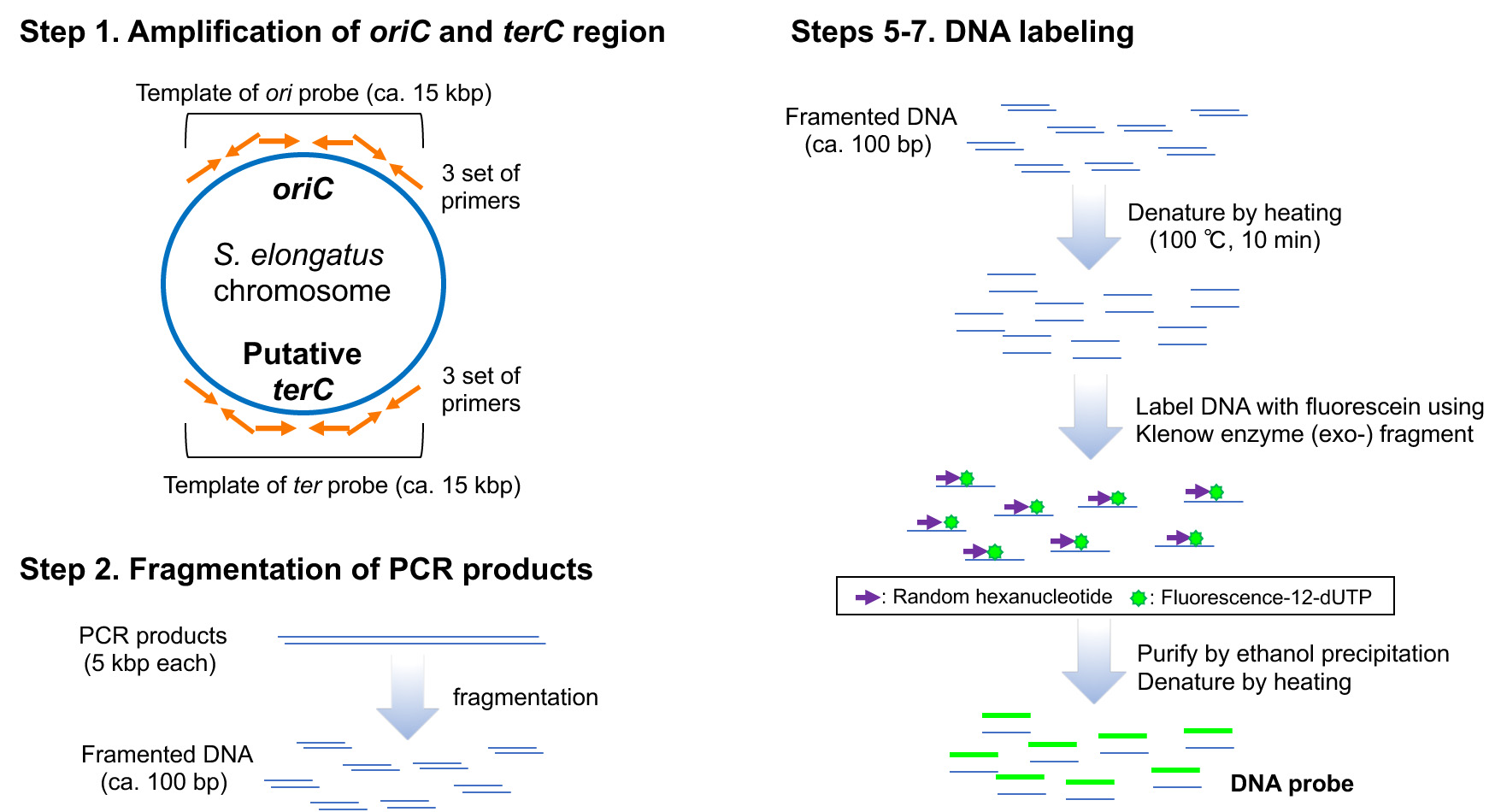
Figure 1. Scheme of preparation of DNA probes
- Preparation of the S. elongatus 7942 culture
- Streak out and pre-incubate S. elongatus 7942 cells on BG-11 agar plates at 30 °C under continuous illumination (40 μE/m2/sec) for 1 week in a plant growth chamber.
- Suspend cells on a BG-11 agar plate in approximately 1 ml of BG-11 medium and inoculate into 80 ml of BG-11 medium in a 100-ml glass tube at an OD750 of approximately 0.1.
- Incubate the culture at 30 °C under continuous illumination (40 μE/m2/sec) and bubble with 2% CO2 in air for 24 h.
- Harvest 5 ml of cell culture and centrifuge at 3,100 x g for 10 min at 4 °C, and then subject the pelleted cells to fixation as described below.
- Fixation
- Suspend the cell pellet in 100 μl medium, add 1 ml chilled fixation solution, and incubate for exactly 5 min at -80 °C.
- Centrifuge the fixed cells at 20,000 x g for 1 min, wash the cell pellet twice with 1 ml of PBS, and store the cell at -30 °C until further analysis.
- Assess the quality of the fixed cells (cell shape, chromosomal morphology and chlorophyll fluorescence) by fluorescence microscopy. For this assessment, mix a few of the fixed cells with 4 μl mounting solution containing DAPI (final conc. 5 μg/ml) and analyze by using an Olympus BX53 fluorescence microscope (Figure 2). In comparison to the untreated cells, the chlorophyll fluorescence of the fixed cells should be decreased. If remarkable reduction of cell size was observed for the fixed cells, shorten the incubation time at -80 °C in Step C1.
Note: The fluorescence of DAPI is observed without incubation. To avoid reduction of fluorescence, we covered the slides with aluminum foil.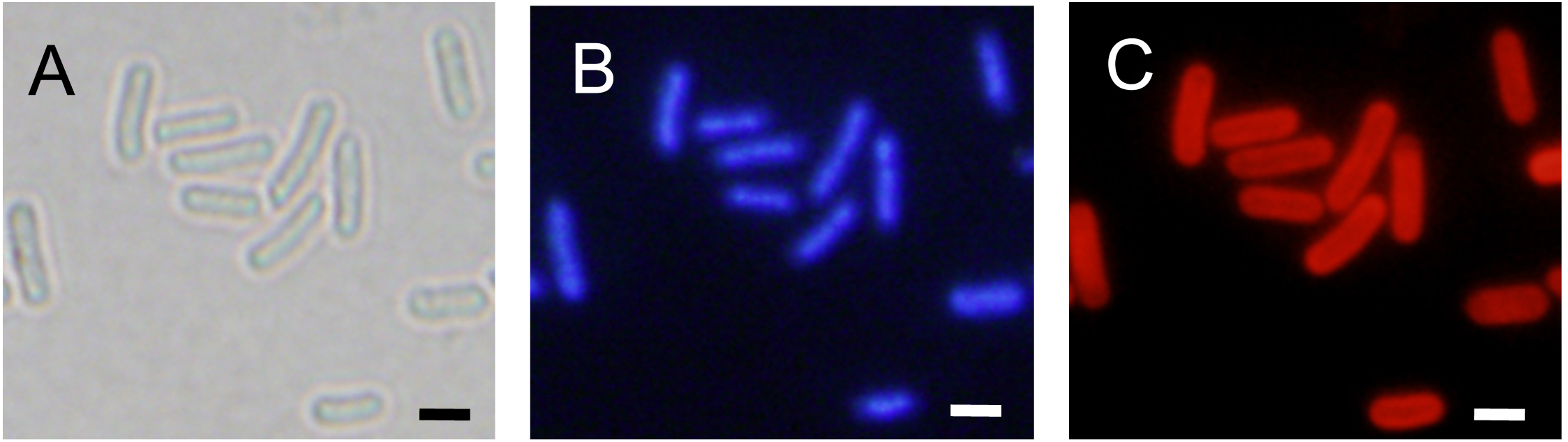
Figure 2. Assessment of cell shape and chromosomal morphology of the fixed cells. The fixed cells were stained with DAPI and examined with fluorescence microscopy. The images shown are as follows: bright-field (A), DAPI staining (B), and chlorophyll fluorescence (C). Scale bars = 2 μm.
- FISH
- Resuspend the fixed cell pellet (Procedure C, Step 2) in 1 ml 0.05% (w/v) Triton X-100 in PBS and incubate for 15 min at room temperature.
- Centrifuge at 20, 000 x g for 1 min and then wash twice with 1 ml of PBS.
- For permeabilization, resuspend the cell pellet in 1 ml of 0.2 mg/ml lysozyme in PBS solution, and incubate for 30 min at 37 °C.
- Centrifuge at 20, 000 x g for 1 min and then wash the cell pellet twice with 1 ml of PBS.
- Spot and spread 10 μl of fixed cell suspension on a poly-L-lysine-coated glass slide with a pipet tip and then let air dry at room temperature.
- For washing the slides, submerge each glass slide into a glass vat filled with water, and remove the slides from the glass vat.
- Incubate each slide in cold (-30 °C) 70% ethanol in a glass vat for 5 min and then wash the slides in another two glass vats (90% and then 100%) each for 5 min at room temperature. Next, air dry the slides.
- Add 5 μl of hybridization solution containing the denatured probe labeled with fluorescence-12-dUTP, as described above on each slide. And then incubate the slides overnight at 42 °C in a humidified chamber (FormaTM direct heat CO2 Incubator with a water-filled vat on the bottom of chamber).
- After hybridization, wash the slides twice with 50% formamide in 1x SSC at 37 °C in a glass vat for 10 min. Wash the slides with 2x and 4x SSC solutions in glass vats at room temperature for 5 min each.
- Air dry the slides at room temperature, and then apply 5 μl of mounting solution to the slide. Seal the slide with a cover glass, and then examine under an Olympus FSX100 fluorescence microscope. Take three images of the cells: DAPI (excitation: 360-370 nm, emission: 420-460 nm), chlorophyll fluorescence (excitation: 530-550 nm, emission: 575 nm), and FISH fluorescence (excitation: 460-495 nm, emission: 510-550 nm) (Figure 3).
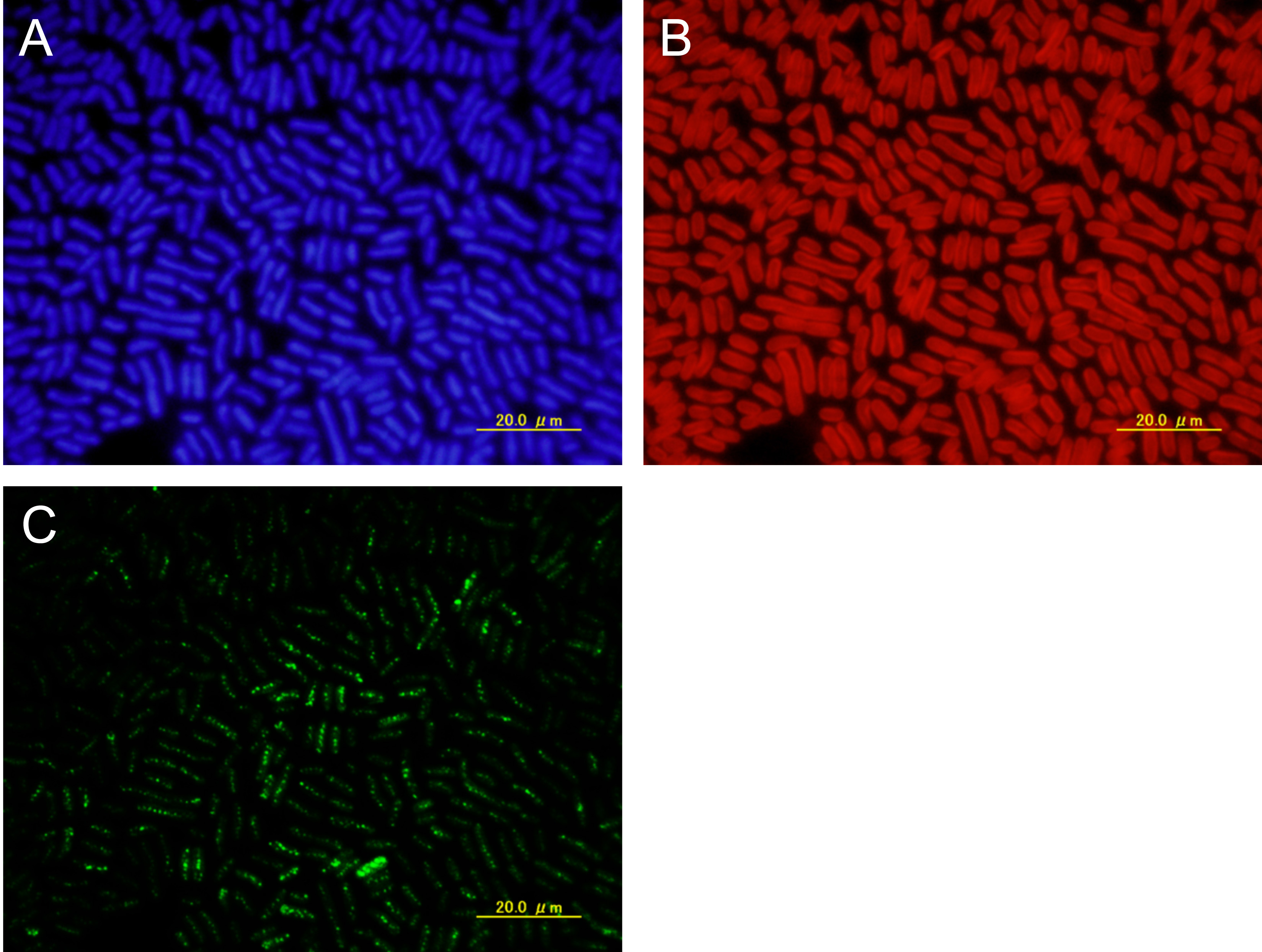
Figure 3. Raw images. Fixed cells were examined by fluorescence in situ hybridization (FISH) using the ori probe, which covered approximately 13 kb encompassing the ori region. Shown are DAPI-stained cells (A), intrinsic chlorophyll fluorescence of cells (B), and FISH foci (C). Scale bars = 20 μm.
Data analysis
- Merge the images with software for the FSX 100 (FSX-BSW).
- Apply changes in brightness and contrast equally to the entire image and to all corresponding images within the same dataset.
- Crop and resize images using Adobe Photoshop, and arrange the images by showing DAPI fluorescence, chlorophyll fluorescence, FISH foci (Figure 4).
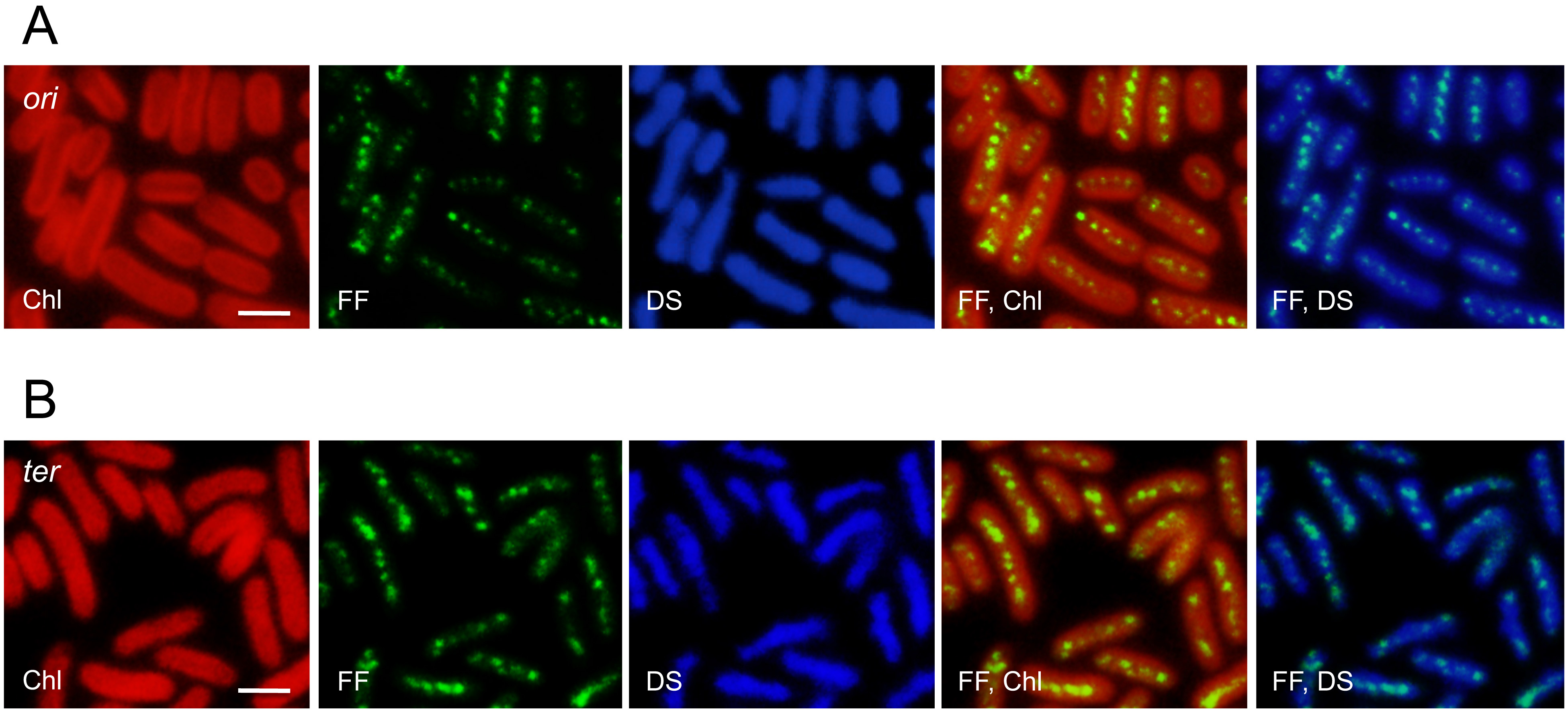
Figure 4. Distribution of the ori region and putative ter region in S. elongatus 7942 cells. Fixed cells were examined by fluorescence in situ hybridization (FISH) using ori (A) and ter probes (B), which covered approximately 13 kb of each region. Shown are the intrinsic chlorophyll fluorescence (Chl), FISH foci (FF), DAPI-stained cells (DS), and merged images. Scale bars = 2 μm.
Recipes
- Hybridization solution (100 ml, store at -20 °C)
Formamide 50 ml 20x SSC 10 ml Salmon sperm DNA 10 mg ddH2O up to 100 ml - Fixation solution (10 ml, prior preparation)
Paraformaldehyde 0.1 g Dimethyl sulfoxide 1 ml 10 N NaOH 20 μl Methanol 8.88 ml - PBS buffer (store at room temperature)
PBS powder 9.6 g ddH2O up to 1 L Autoclave (A. C.) (120 °C, 20 min) - Lysozyme solution (10 ml, prior preparation)
Lysozyme 2 mg 1 M Tris-HCl (pH 7.5) 250 μl 0.5 M EDTA 200 μl PBS 9.55 ml - 20x Standard Saline Citrate (SSC) (1 L, store at room temperature)
A. C. (120 °C, 20 min)NaCl 175.32 g C6H5Na3O7•2H2O 88.23 g pH adjusted to 7.0 with HCl ddH2O up to 1 L - Mounting solution (1 ml, store at 4 °C)
DAPI (0.2 mg/ml) 25 μl PBS 975 μl
Acknowledgments
This protocol was adapted from Watanabe et al. (2018). This work was supported in part by Grants-in-Aid 25850056 and 17H05451 from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.W. The authors declare no conflicts of interests.
References
- Binder, B. J. and Chisholm, S. W. (1995). Cell cycle regulation in marine Synechococcus sp. strains. Appl Environ Microbiol 61(2): 708-717.
- Castenholz, R. W. (1988). Culturing methods for cyanobacteria. Meth Enzymol 167: 68-93.
- Chen, A. H., Afonso, B., Silver, P. A. and Savage, D. F. (2012). Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PLoS One 7(10): e47837.
- Griese, M., Lange, C. and Soppa, J. (2011). Ploidy in cyanobacteria. FEMS Microbiol Lett 323(2): 124-131.
- Jain, I. H., Vijayan, V. and O'Shea, E. K. (2012). Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci U S A 109(34): 13638-13643.
- Labarre, J., Chauvat, F. and Thuriaux, P. (1989). Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J Bacteriol 171(6): 3449-3457.
- Mann, N. and Carr, N. G. (1974). Control of macromolecular composition and cell division in the blue-green algae Anacystis nidulans. J Gen Microbiol 83(2): 399-405.
- Watanabe, S., Noda, A., Ohbayashi, R., Uchioke, K., Kurihara, A., Nakatake, S., Morioka, S., Kanesaki, Y., Chibazakura, T. and Yoshikawa, H. (2018). ParA-like protein influences the distribution of multi-copy chromosomes in cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 164(1): 45-56.
- Watanabe, S., Ohbayashi, R., Kanesaki, Y., Saito, N., Chibazakura, T., Soga, T. and Yoshikawa, H. (2015). Intensive DNA replication and metabolism during the lag phase in cyanobacteria. PLoS One 10(9): e0136800.
- Watanabe, S., Ohbayashi, R., Shiwa, Y., Noda, A., Kanesaki, Y., Chibazakura, T. and Yoshikawa, H. (2012). Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol Microbiol 83(4): 856-865.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ohbayashi, R., Yoshikawa, H. and Watanabe, S. (2018). Direct Visualization of the Multicopy Chromosomes in Cyanobacterium Synechococcus elongatus PCC 7942. Bio-protocol 8(15): e2958. DOI: 10.21769/BioProtoc.2958.
Category
Microbiology > Microbial cell biology > Cell staining
Molecular Biology > DNA > DNA labeling
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











