- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
HCV Reporter System (Viral Infection-Activated Split-Intein-Mediated Reporter System) for Testing Virus Cell-to-cell Transmission ex-vivo
Published: Vol 8, Iss 15, Aug 5, 2018 DOI: 10.21769/BioProtoc.2949 Views: 6427
Reviewed by: David PaulJose Antonio Reyes-DariasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Unbiased Screening of Activated Receptor Tyrosine Kinases (RTKs) in Tumor Extracts Using a Mouse Phospho-RTK Array Kit
Julian Naipauer [...] Enrique A. Mesri
Apr 20, 2019 5678 Views

Generation and Implementation of Reporter BHK-21 Cells for Live Imaging of Flavivirus Infection
Jorge L. Arias-Arias and Rodrigo Mora-Rodríguez
Mar 5, 2021 4367 Views

Selection of Vaccinia Virus Recombinants Using CRISPR/Cas9
Anjali Gowripalan [...] David C. Tscharke
Dec 20, 2021 3416 Views
Abstract
Hepatitis C virus (HCV) spread involves two distinct entry pathways: cell-free transmission and cell-to-cell transmission. Cell-to-cell transmission is not only an efficient way for viruses to spread but also an effective method for escaping neutralizing antibodies. We adapted the viral infection-activated split-intein-mediated reporter system (VISI) and developed a straightforward model for Live-cell monitoring of HCV cell-to-cell transmission ex-vivo: co-culture of HCV infected donor cells (red signal) with uninfected recipient cells (green signal) and elimination of the cell-free transmission by adding potent neutralizing antibody AR3A in the supernatant. With this model, the efficiency of cell-to-cell transmission can be evaluated by counting the number of foci designated by the green signal of recipient cells.
Keywords: HCVBackground
Accumulating evidence support that viruses can use different routes of spread in infected tissues (Sattentau, 2008; Zhong et al., 2013). For HCV transmission, both cell-free transmission and cell-to-cell transmission can mediate virus transfer between hepatocytes. While cell-free transmission initiates HCV infection, cell-cell transmission is thought to transfer HCV to adjacent hepatocytes directly. It provides an excellent way to resist the neutralizing antibodies and contribute to the viral persistence (Brimacombe et al., 2011; Xiao et al., 2014). Previous articles also proved some host factors which contributed to cell-cell transmission, such as scavenger receptor BI (SR-BI), CD81, tight junction proteins claudin-1 (CLDN1), Occludin (OCLN), epidermal growth factor receptor (EGFR) (Witteveldt et al., 2009; Catanese et al., 2013; Zona et al., 2013). But the exact mechanisms of this process still need to explore. We optimized a viral infection-activated split-intein-mediated reporter system (VISI) for live-cell visualization of HCV infection (Zhao et al., 2017). Based on the study in Huh7.5.1 cell line using a technique of split GFP/RFP reconstitution by intein protein splicing (Figure 1A), it showed that VISI system is a very sensitive and low background system. With this system, we can clearly visualize the HCV infected cell by its nuclear fluorescence signal. In addition, combining VISI-GFP and VISI-mCherry cells, we can further monitor HCV cell-to-cell transmission in the presence of potent neutralizing antibody AR3A.
Materials and Reagents
- For cell culture materials
- Sterile 100 mm polystyrene Petri dish (Thermo Fisher Scientific, catalog number: 172931 )
- Sterile flat-bottom 96-well plate (Thermo Fisher Scientific, catalog number: 167008 )
- Sterile 60 mm polystyrene Petri dish (Thermo Fisher Scientific, catalog number: 150288 )
- Sterile 6-well plate (Thermo Fisher Scientific, catalog number: 140675 )
- Sterile 50 ml Conical Centrifuge Tube (Thermo Fisher Scientific, catalog number: 339652 )
- Sterile 100 mm polystyrene Petri dish (Thermo Fisher Scientific, catalog number: 172931 )
- Plasmids and Cell lines
- Lentiviral vector:
pWPI-blasticidin-NLS-GFPn-INTEINn (Genbank: KY067203)
pWPI-puromycin-INTEINc-GFPc-NLS-IPS (Genbank: KY067204) (for construction of Huh7.5.1-VISI-GFP cell line)
pWPI-blasticidin-NLS-mCherry(n)-INTEINn (Genbank: KY067205)
pWPI-puromycin-INTEINc-mCherry(c)-NLS-IPS (Genbank: KY067206) (for construction of Huh7.5.1-VISI-mCherry cell line) - Two auxiliary plasmids:
psPAX2 (the HIV-1 packaging plasmid)
pMD2.G (a vesicular stomatitis virus glycoprotein [SV-G] expression vector)
- 293T cell (ATCC, catalog number: CRL-3216 )
- Huh7.5.1 cell (Human hepatocyte-derived cell line Huh7.5.1 is kindly provided by professor F. Chisari)
- Lentiviral vector:
- HCV virus strains
- HCV JC1 (GenBank: JF343782.1)
- Subgenomic JFH1 (sgJFH1) (GenBank: AB114136.1)
- HCV JC1 (GenBank: JF343782.1)
- For cell culture medium
- 1x phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010049 )
- 0.25% Trypsin (Thermo Fisher Scientific, GibcoTM, catalog number: 25200072 )
- DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: C11965500CP )
- Fetal bovine serum (Thermo Fisher Scientific, GibcoTM, catalog number: 10099141 )
- 100x Nonessential amino acids (Thermo Fisher Scientific, GibcoTM, catalog number: 11140050 )
- 100x Penicillin/streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Complete DMEM (see Recipes)
- 1x phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010049 )
- For transcription in vitro
- MEGAscript® T7 Transcription Kit (Thermo Fisher Scientific, catalog number: AM1333 )
- MEGAscript® T7 Transcription Kit (Thermo Fisher Scientific, catalog number: AM1333 )
- For electroporation buffer
- ATP (Thermo Fisher Scientific, catalog number: R0441 )
- L-Glutathione (Sigma-Aldrich, catalog number: V900456-25G )
- Potassium chloride (KCl) (Sinopharm Chemical Reagent, catalog number: 10016318 )
- Calcium chloride (CaCl2) (Shanghai Experiment Reagent, catalog number: 117600 )
- Dipotassium hydrogen phosphate (K2HPO4) (Shanghai Experiment Reagent, catalog number: 168120 )
- Potassium phosphate monobasic (KH2PO4) (Shanghai Experiment Reagent, catalog number: 175650 )
- HEPES (Sigma-Aldrich, catalog number: H4034-25G )
- EGTA (Sangon Biotech, catalog number: E0732-50G )
- Magnesium chloride (MgCl2) (Sinopharm Chemical Reagent, catalog number: 10012818 )
- Cytomix buffer (see Recipes)
- ATP (Thermo Fisher Scientific, catalog number: R0441 )
- For Calcium Phosphate (CaPO4) transfection buffer
- HEPES (Sigma-Aldrich, catalog number: H4034-25G )
- Sodium chloride (NaCl) (Sinopharm Chemical Reagent, catalog number: 10019318 )
- Disodium hydrogen phosphate (Na2HPO4·12H2O) (Shanghai Experiment Reagent, catalog number: 174710 )
- Calcium chloride (CaCl2) (Shanghai Experiment Reagent, catalog number: 117600 )
- Calcium Phosphate (CaPO4) transfection buffer (see Recipes)
- HEPES (Sigma-Aldrich, catalog number: H4034-25G )
- HCV-neutralizing antibody
- Antibody AR3A
Note: AR3A is kindly provided by professor M. Law.
- Antibody AR3A
- For cell line selecting
- Puromycin (Sigma-Aldrich, catalog number: P8833-25MG )
- Blasticidin (Thermo Fisher Scientific, GibcoTM, catalog number: R21001 )
- Puromycin (Sigma-Aldrich, catalog number: P8833-25MG )
Equipment
- Electroporator (Bio-Rad Laboratories, model: Gene Pulser XcellTM )
- Electroporation cuvette (Bio-Rad Laboratories, Gene Pulser cuvette, 0.4 cm)
- Fluorescence microscope (Olympus, model: IX53 )
Software
- GraphPad Prism (GraphPad Software, https://www.graphpad.com/)
Procedure
- Construction of HCV reporter system (VISI system) cell line
The mechanism and construction of VISI system have been described in detail in our research paper (Zhao et al., 2017). The materials (plasmids and cell lines used in VISI system) are available upon request from our lab.- Plasmid construction
Technically, most reporters are applicable to VISI system, such as green fluorescent protein (GFP), red fluorescent protein (RFP), luciferases, antibiotic-resistant proteins. Here, we chose two different fluorescent proteins: green fluorescent protein (GFP, green signal) and mCherry (red signal), for HCV reporter system construction (VISI-GFP/VISI-mCherry). N/C-terminal sequences of VISI are inserted into lentiviral vectors pWPI-blasticidin or pWPI-puromycin, respectively.
Note: These four DNA sequences have been uploaded (Genbank: KY067203 [N-terminal of VISI-GFP], KY067204 [C-terminal of VISI-GFP], KY067205 [N-terminal of VISI-mCherry], KY067206 [C-terminal of VISI-mCherry]). - Generation of Huh7.5.1-VISI by lentiviral transduction
- Lentiviral production
Day 1:
Plate 1.2 x 106 293T cells in complete DMEM into a 6 cm dish.
Day 2:
Replace the medium with 4 ml fresh complete DMEM and warm up the solutions of calcium phosphate (CaPO4) transfection (2x HEPES solution, 2.5 M calcium chloride solution, sterile H2O). After 1-2 h, dilute 15 μg DNA (pWPI vector:psPAX2:pMD2.G = 6.4 μg:6.4 μg:2.1 μg) in a final volume of 270 μl sterile H2O, and add 30 μl calcium chloride solution (2.5 M). Then vortex the mixed liquid gently and add 2x HEPES solution (300 μl) drop by drop simultaneously. Finally transfer the mixture into a 6 cm dish by dropwise, gently shake to distribute the precipitate evenly.
Day 3:
Change the medium with 4 ml fresh complete DMEM at 16 h post calcium phosphate transfection.
Day 4:
Harvest the supernatant (lentivirus) at 48 h post calcium phosphate transfection, after filtration with 0.45 μm filter, store in aliquots at -80 °C. - These lentiviral preparations are used to transduce the targeted cell line Huh7.5.1 of which 5 x 105 cells are seeded into a 6-well plate. Eight hours after transduction, discard the lentiviral preparation and replace with fresh complete medium. Two days later, resuspend the transduced cells and transfer them into a 10 cm dish. Then add antibiotics after cell adhesion.
- After selection by both puromycin (1 μg/ml) and blasticidin (4 μg/ml) for days, Huh7.5.1-VISI-GFP and Huh7.5.1-VISI-mCherry cell lines will be successfully obtained, respectively.
- Lentiviral production
- Plasmid construction
- Construction of cell model for testing cell-to-cell transmission
We choose Huh7.5.1-VISI-mCherry as donor cells that were electroporated with purified HCV RNA. Huh7.5.1-VISI-GFP is used as recipient cells which could receive virus genome from donor cells.- First, Huh7.5.1-VISI-mCherry is electroporated with in vitro transcripts of HCV (Jc1) or subgenomic JFH1 (sgJFH1) which in vitro transcripted from HCV infectious clone pFK-JC1 or subgenomic construct pFK-sgJFH1 (Long et al., 2011). These two HCV genomes could be the positive and negative control of this experiment.
- pFK-JC1 and pFK-sgJFH1 are linearized by restriction enzyme Mlu1 or Xba1 respectively.
- Assemble transcription reaction at room temperature (1 μg linearized template, 2 μl Enzyme Mix, 2 μl ATP solution, 2 μl CTP solution, 2 μl GTP solution, 2 μl UTP solution, 2 μl 10x Reaction buffer, Add proper amount of Nuclease-free water to 20 μl), Mix thoroughly and incubate for 3 h at 37 °C.
- Then add 1 μl DNase to digest the template and incubate for 15 min at 37 °C.
- Lithium Chloride (LiCl) precipitation can be used to purified transcriptional RNA. Add 30 μl LiCl precipitation solution and chill for 1 h at -20 °C, then centrifuge for 15 min at maximum speed to pellet the RNA.
- After wash twice with 70% ethanol, resuspend the RNA pellet with 20 μl RNase-free water. And store in 10 μg aliquots at -80 °C.
- Add 10 μg of in vitro transcripts (RNA) with the cell suspension–5 x 106 Huh7.5.1-VISI-mCherry cells are resuspended in 400 μl cytomix buffer (Recipe 2). Then the cells are transfected by electroporation at 960 mF and 270 V using Bio-Rad Gene Pulser system and a cuvette with a gap width of 0.4 cm (Bio-Rad). Immediately after electroporation, resuspend the cells in complete DMEM and seed as required.
- pFK-JC1 and pFK-sgJFH1 are linearized by restriction enzyme Mlu1 or Xba1 respectively.
- Electroporated Huh7.5.1-VISI-mCherry are individualized and mixed with Huh7.5.1-VISI-GFP cells at a ratio of 1:30.
Note: For single well of 96-well plates, it needs 1,000 Huh7.5.1-VISI-mCherry cells plus 30,000 Huh7.5.1-VISI-GFP cells. - Culture the cells in the presence of HCV-neutralizing antibody AR3A at 50 μg/ml to block cell-free transmission (Figure 1B). Replace antibody-containing medium every 24 h.
- After co-culture for 72-96 h, HCV cell-to-cell transmission efficiency can be evaluated by capturing the fluorescent images and examining the number of GFP or mCherry glowing cells.
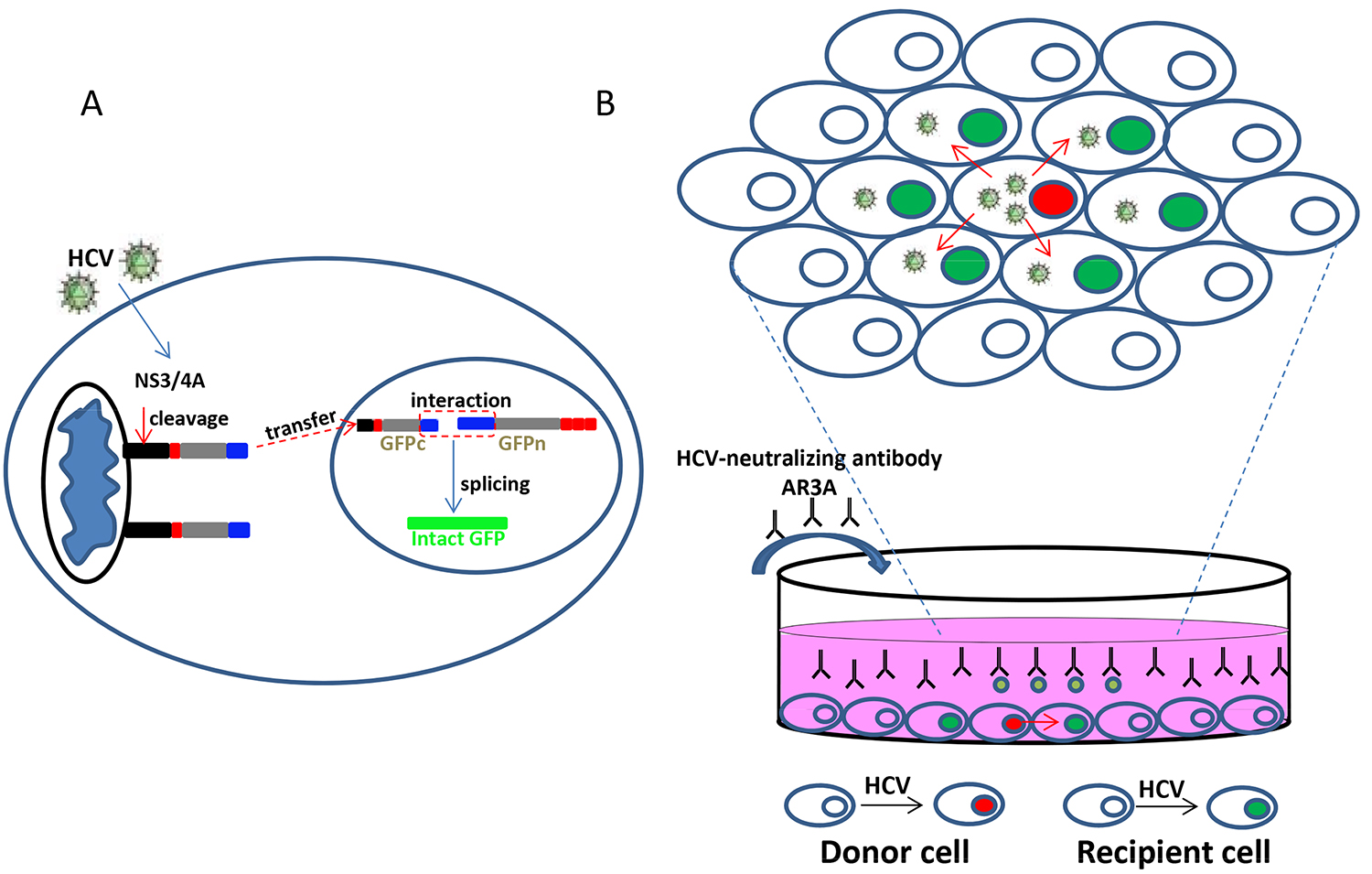
Figure 1. Schematic diagram of VISI system and HCV cell-to-cell transmission system. A. Fusion proteins of N-terminal and C-terminal halves of VISI are located in nucleus and mitochondria by different location signal (nuclear localization signal [NLS] or IPS), respectively; upon virus infection, HCV NS3-4A cleavage activates transportation of the C-terminal piece from mitochondria to the nucleus where intact GFP is reconstituted through split-intein splicing. B. Huh7.5.1-VISI-mCherry cells electroporated with the HCV genome are used as donor cells; and Huh7.5.1-VISI-GFP cells are used as recipient cells. Donor cells are co-cultured with recipient cells at a cell number ratio of 1:30. To block cell-free transmission, we added HCV-neutralizing antibody AR3A into the supernatant. After incubation for 96 h, images are taken under a fluorescence microscope (Figure 2A).
- First, Huh7.5.1-VISI-mCherry is electroporated with in vitro transcripts of HCV (Jc1) or subgenomic JFH1 (sgJFH1) which in vitro transcripted from HCV infectious clone pFK-JC1 or subgenomic construct pFK-sgJFH1 (Long et al., 2011). These two HCV genomes could be the positive and negative control of this experiment.
Data analysis
- For experimental design, there are three replicates for each experiment. For each replicate, we capture 10-15 fluorescent images so as to count a minimum of 40 virus foci (Figure 2A).
- For data analysis, to evaluate the efficiency of HCV cell-to-cell transmission, we randomly take the fluorescent images of virus foci that several red donor cells are surrounded by some green recipient cells or not (Figure 2A). The numbers of red donor cells and green recipient cells should be counted, respectively (Figure 2B).
- For statistical analysis, we use GraphPad Prism to analyze the difference between samples by using the two-tailed, unpaired Student’s t-test.
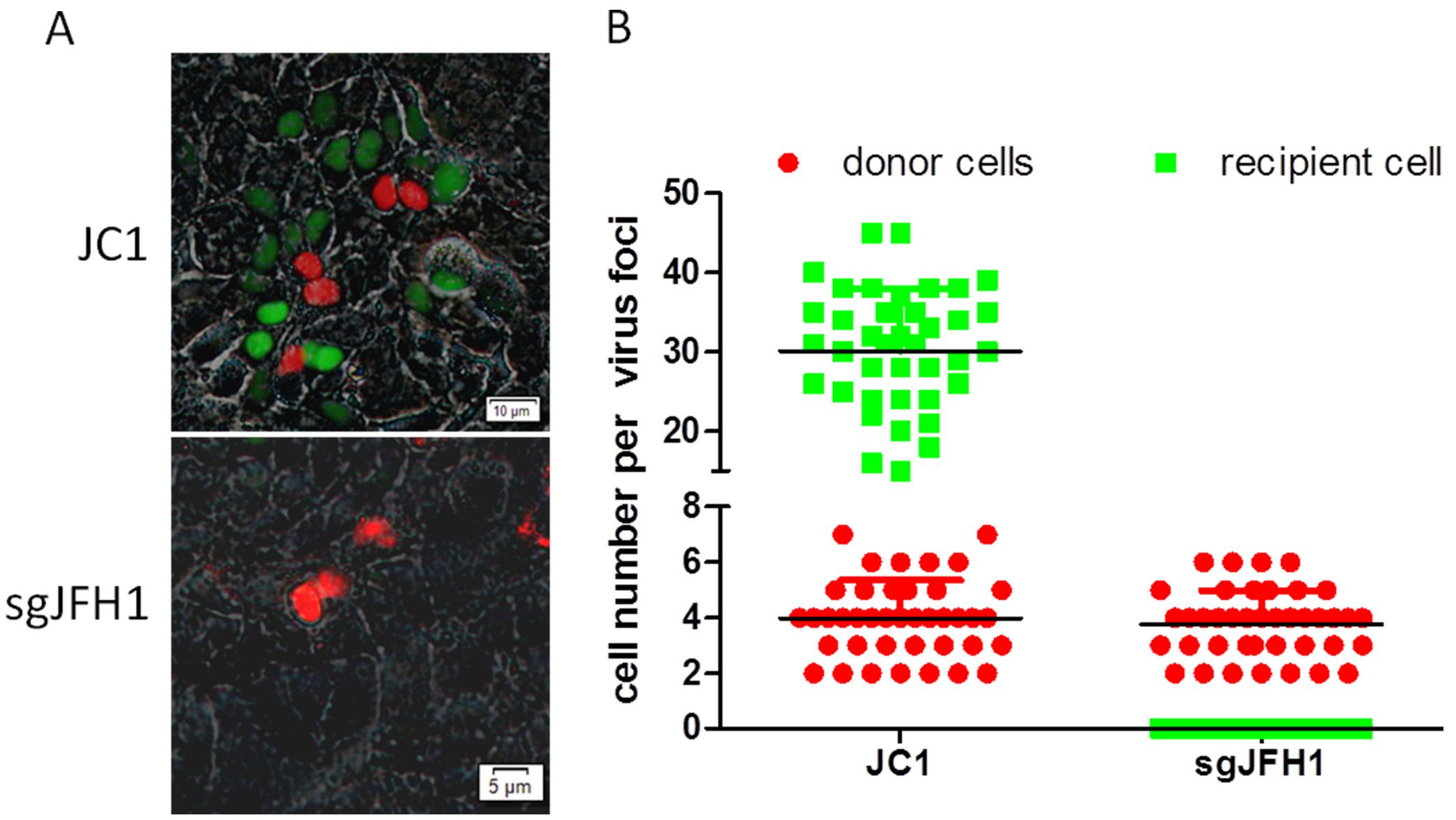
Figure 2. Data processing and analysis of HCV cell-to-cell transmission system. A. HCV cell-to-cell transmission phenomena is observed by fluorescence microscopy after co-culture for 96 h. Donor cells Huh7.5.1-VISI-mCherry electroporated with the HCV genome JC1 or subgenomic JFH1 (sgJFH1), and Huh7.5.1-VISI-GFP cells are used as recipient cells. So red nuclear fluorescence represent HCV positive donor cells and green nuclear fluorescence represent recipient cells which infected with HCV in cell-to-cell transmission. B. Numbers of HCV-positive donor cells (Huh7.5.1-VISI-mCherry cells) or HCV-positive recipient cells (Huh7.5.1-VISI-GFP cells) per focus after co-culture for 96 h are counted. In the scatter plot, red circles represent the number of HCV-positive donor cells per focus; green squares represent the number of HCV-positive recipient cells per virus foci; horizontal lines represent the median for 40 randomly selected foci.
Recipes
- Complete DMEM
DMEM medium mix with 10% FBS (440 ml DMEM + 50 ml FBS)
1x Non-essential amino acids (5 ml)
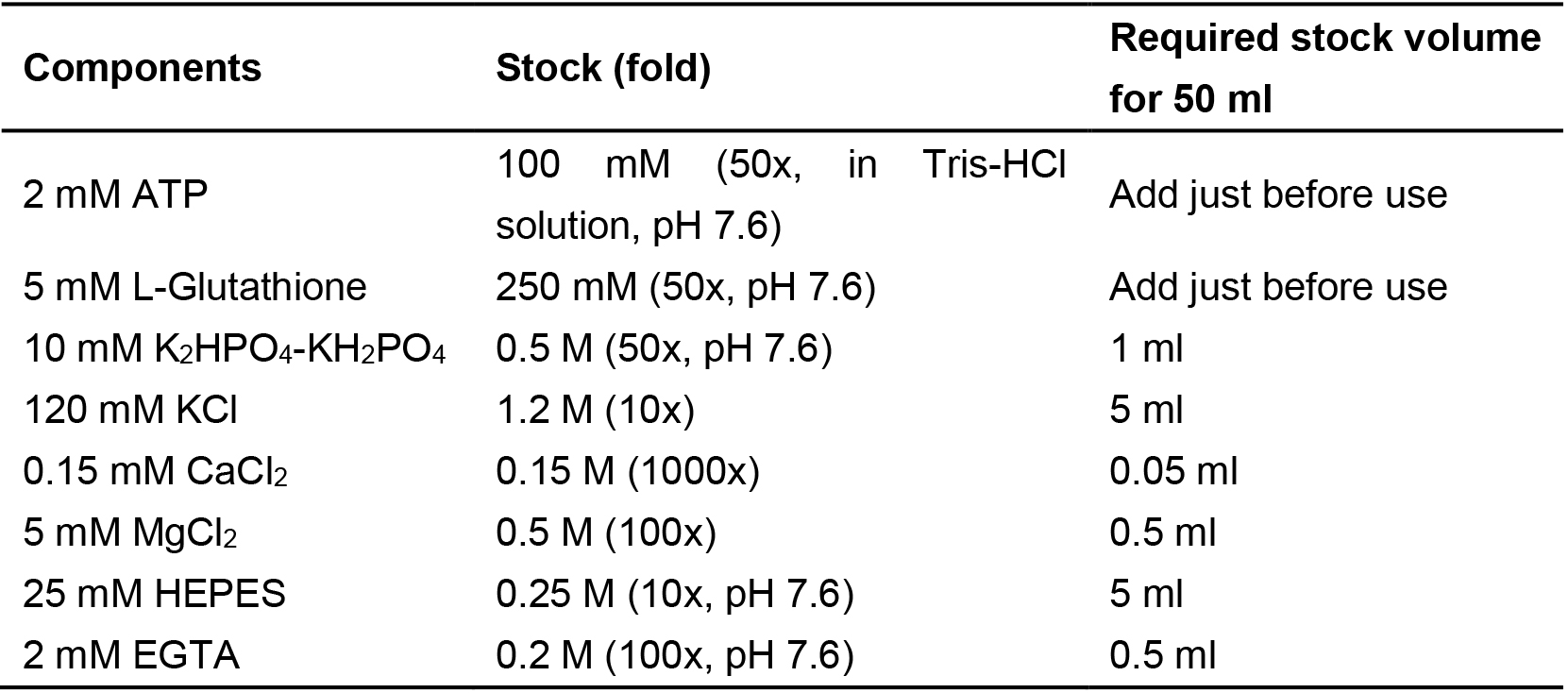
1x Penicillin/streptomycin (5 ml Penicillin/streptomycin) - Cytomix buffer, pH 7.6 (electroporation buffer)
Notes:- The pH value of the mixture solution is adjusted to 7.6 with 3 M potassium hydrate (KOH). Filtrate through a 0.22 μm filter, and store in 10 ml aliquots at -20 °C.
- The above solutions are prepared with double distilled water unless otherwise indicated.
- The pH value of the mixture solution is adjusted to 7.6 with 3 M potassium hydrate (KOH). Filtrate through a 0.22 μm filter, and store in 10 ml aliquots at -20 °C.
- Calcium Phosphate (CaPO4) transfection buffer
- 2.5 M CaCl2 (in ddH2O), filtrate through a 0.22 μm filter, and store in 10 ml aliquots at -20 °C
- 2x HEPES buffer, pH 7.1
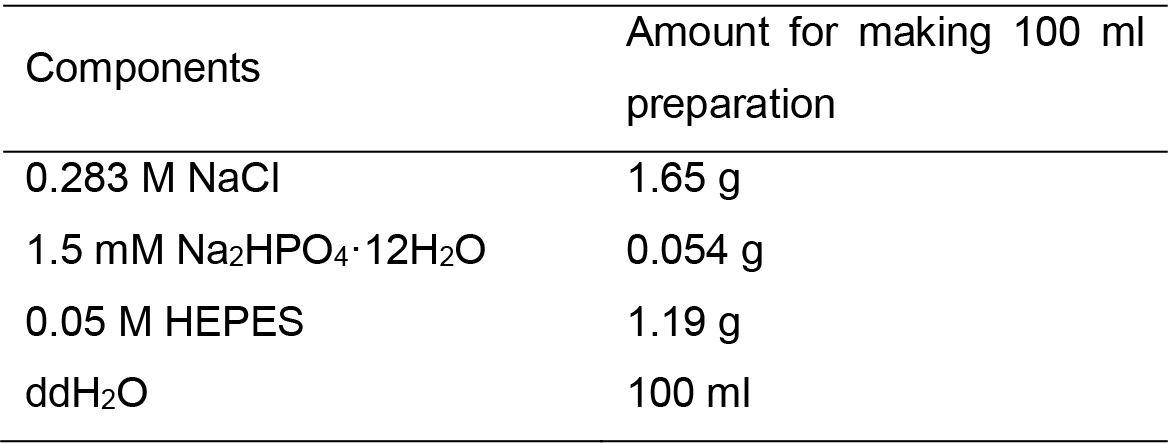
Note: The pH value of the mixture solution is adjusted to 7.1. Filtrate through a 0.22 μm filter, and store in 10 ml aliquots at -20 °C. For single calcium phosphate (CaPO4) transfection in a 6-cm dish, it needs 270 μl ddH2O (contain 15 μg DNA plasmids), 30 μl 2.5 M Cacl2 and 300 μl 2x HEPES buffer.
- 2.5 M CaCl2 (in ddH2O), filtrate through a 0.22 μm filter, and store in 10 ml aliquots at -20 °C
Acknowledgments
This protocol was adapted from procedures published in Zhao et al. (2017). We thank F. Chisari for kindly providing Huh7.5.1 cell line; M. Law for the gift of HCV-neutralizing antibody AR3A; and T. Wakita, C. M. Rice, J. Bukh, and R. Bartenschlager for providing HCV strains.
This work was supported by the National Key R&D program of China (2016YFC1200400), the “100 talents program” from the Chinese Academy of Sciences, and the National Science and Technology Major Project of the Ministry of Science and Technology of China (2014ZX10002002-001-004 and 2015CB554300). The authors declare that there are no conflicts of interest.
References
- Brimacombe, C. L., Grove, J., Meredith, L. W., Hu, K., Syder, A. J., Flores, M. V., Timpe, J. M., Krieger, S. E., Baumert, T. F., Tellinghuisen, T. L., Wong-Staal, F., Balfe, P. and McKeating, J. A. (2011). Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol 85(1): 596-605.
- Catanese, M. T., Loureiro, J., Jones, C. T., Dorner, M., von Hahn, T. and Rice, C. M. (2013). Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol 87(15): 8282-8293.
- Long, G., Hiet, M. S., Windisch, M. P., Lee, J. Y., Lohmann, V. and Bartenschlager, R. (2011). Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology 141(3): 1057-1066.
- Sattentau, Q. (2008). Avoiding the void: cell-to-cell spread of human viruses. Nat Rev Microbiol 6(11): 815-826.
- Witteveldt, J., Evans, M. J., Bitzegeio, J., Koutsoudakis, G., Owsianka, A. M., Angus, A. G., Keck, Z. Y., Foung, S. K., Pietschmann, T., Rice, C. M. and Patel, A. H. (2009). CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol 90: 48-58.
- Xiao, F., Fofana, I., Heydmann, L., Barth, H., Soulier, E., Habersetzer, F., Doffoel, M., Bukh, J., Patel, A. H., Zeisel, M. B. and Baumert, T. F. (2014). Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog 10(5): e1004128.
- Zhao, F., Zhao, T., Deng, L., Lv, D., Zhang, X., Pan, X., Xu, J. and Long, G. (2017). Visualizing the essential role of complete virion assembly machinery in efficient hepatitis C virus cell-to-cell transmission by a viral infection-activated split-intein-mediated reporter system. J Virol 91(2): e01720-16.
- Zhong, P., Agosto, L. M., Munro, J. B. and Mothes, W. (2013). Cell-to-cell transmission of viruses. Curr Opin Virol 3(1): 44-50.
- Zona, L., Lupberger, J., Sidahmed-Adrar, N., Thumann, C., Harris, H. J., Barnes, A., Florentin, J., Tawar, R. G., Xiao, F., Turek, M., Durand, S. C., Duong, F. H., Heim, M. H., Cosset, F. L., Hirsch, I., Samuel, D., Brino, L., Zeisel, M. B., Le Naour, F., McKeating, J. A. and Baumert, T. F. (2013). HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 13(3): 302-313.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhao, F., Zhao, T., Deng, L., Lv, D., Zhang, X., Pan, X., Xu, J. and Long, G. (2018). HCV Reporter System (Viral Infection-Activated Split-Intein-Mediated Reporter System) for Testing Virus Cell-to-cell Transmission ex-vivo. Bio-protocol 8(15): e2949. DOI: 10.21769/BioProtoc.2949.
Category
Microbiology > in vivo model > Viruses
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link